Machine Learning Quantification of Tumor-Stroma Ratio in Early Muscle Invasive Urothelial Carcinomas
Article Information
Vrabie Camelia D1*, Gangal Marius D2
1Pathology Department, “Sfântul Ioan” Clinical Emergency Hospital, 042122 Bucharest, Romania
2Medical Data Analytics (MEDACS), Rigaud, J0P1P0, PQ, Canada
*Corresponding author: Vrabie Camelia D, Pathology Department, “Sfântul Ioan” Clinical Emergency Hospital, 042122 Bucharest, Romania.
Received: 13 September 2022; Accepted: 19 September 2022; Published: 10 October 2022
Citation: Vrabie Camelia D, Gangal Marius D. Machine Learning Quantification of Tumor-Stroma Ratio in Early Muscle Invasive Urothelial Carcinomas. Journal of Biotechnology and Biomedicine 5 (2022): 185-195.
View / Download Pdf Share at FacebookAbstract
Tumor-Stroma Ratio, a marker of tumor microenvironment, proved to be a reliable independent prognostic predictor in many solid tumors but it’s value in transitional carcinoma is still under research. Visual quantification of tumoral and stromal areas is possible but is time consuming and subjective. Machine learning image segmentation can improve diagnostic precision. Our research interest is to evaluate how precision pathology tools (machine learning segmentation of whole slide images) may improve quantification of the tumor-stroma ratio in early muscle invasive bladder tumors and increase histologic diagnostic prognostic value. 10 cases of pathology stage T2A bladder cancers whole slide images were carefully matched (sex, age and smoking status) with 10 cases of pT2B form the same open database (Cancer Genome Atlas Urothelial Bladder Carcinoma dataset, TCGA-BLCA). The machine learning segmentation used a trained approach and was performed under 3 labels (tumor, stroma, other). The mean tumor to stroma ratio was significant higher (tumor>stroma) in pT2A cohort (p<0.0001). Vital prognostic was different between groups: 90% of subjects were alive at 3 years after diagnostic in pT2A cohort and only 40% in pT2B cohort. Our proof-of-concept study suggest the utility of the tumor-stroma ratio in differentiating challenging diagnostics of early muscle invasive urothelial carcinoma. A larger, real world data study will have to confirm the benefits of this marker in everyday clinical settings.
Keywords
Tumor-Stroma Ratio; Muscle Invasive Bladder Cancer; Tumor Grading; Machine Learning Segmentation; Whole Slide Images
Tumor-Stroma Ratio articles Tumor-Stroma Ratio Research articles Tumor-Stroma Ratio review articles Tumor-Stroma Ratio PubMed articles Tumor-Stroma Ratio PubMed Central articles Tumor-Stroma Ratio 2023 articles Tumor-Stroma Ratio 2024 articles Tumor-Stroma Ratio Scopus articles Tumor-Stroma Ratio impact factor journals Tumor-Stroma Ratio Scopus journals Tumor-Stroma Ratio PubMed journals Tumor-Stroma Ratio medical journals Tumor-Stroma Ratio free journals Tumor-Stroma Ratio best journals Tumor-Stroma Ratio top journals Tumor-Stroma Ratio free medical journals Tumor-Stroma Ratio famous journals Tumor-Stroma Ratio Google Scholar indexed journals Muscle Invasive Bladder Cancer articles Muscle Invasive Bladder Cancer Research articles Muscle Invasive Bladder Cancer review articles Muscle Invasive Bladder Cancer PubMed articles Muscle Invasive Bladder Cancer PubMed Central articles Muscle Invasive Bladder Cancer 2023 articles Muscle Invasive Bladder Cancer 2024 articles Muscle Invasive Bladder Cancer Scopus articles Muscle Invasive Bladder Cancer impact factor journals Muscle Invasive Bladder Cancer Scopus journals Muscle Invasive Bladder Cancer PubMed journals Muscle Invasive Bladder Cancer medical journals Muscle Invasive Bladder Cancer free journals Muscle Invasive Bladder Cancer best journals Muscle Invasive Bladder Cancer top journals Muscle Invasive Bladder Cancer free medical journals Muscle Invasive Bladder Cancer famous journals Muscle Invasive Bladder Cancer Google Scholar indexed journals Tumor Grading articles Tumor Grading Research articles Tumor Grading review articles Tumor Grading PubMed articles Tumor Grading PubMed Central articles Tumor Grading 2023 articles Tumor Grading 2024 articles Tumor Grading Scopus articles Tumor Grading impact factor journals Tumor Grading Scopus journals Tumor Grading PubMed journals Tumor Grading medical journals Tumor Grading free journals Tumor Grading best journals Tumor Grading top journals Tumor Grading free medical journals Tumor Grading famous journals Tumor Grading Google Scholar indexed journals Machine Learning Segmentation articles Machine Learning Segmentation Research articles Machine Learning Segmentation review articles Machine Learning Segmentation PubMed articles Machine Learning Segmentation PubMed Central articles Machine Learning Segmentation 2023 articles Machine Learning Segmentation 2024 articles Machine Learning Segmentation Scopus articles Machine Learning Segmentation impact factor journals Machine Learning Segmentation Scopus journals Machine Learning Segmentation PubMed journals Machine Learning Segmentation medical journals Machine Learning Segmentation free journals Machine Learning Segmentation best journals Machine Learning Segmentation top journals Machine Learning Segmentation free medical journals Machine Learning Segmentation famous journals Machine Learning Segmentation Google Scholar indexed journals Whole Slide Images articles Whole Slide Images Research articles Whole Slide Images review articles Whole Slide Images PubMed articles Whole Slide Images PubMed Central articles Whole Slide Images 2023 articles Whole Slide Images 2024 articles Whole Slide Images Scopus articles Whole Slide Images impact factor journals Whole Slide Images Scopus journals Whole Slide Images PubMed journals Whole Slide Images medical journals Whole Slide Images free journals Whole Slide Images best journals Whole Slide Images top journals Whole Slide Images free medical journals Whole Slide Images famous journals Whole Slide Images Google Scholar indexed journals urinary malignancy articles urinary malignancy Research articles urinary malignancy review articles urinary malignancy PubMed articles urinary malignancy PubMed Central articles urinary malignancy 2023 articles urinary malignancy 2024 articles urinary malignancy Scopus articles urinary malignancy impact factor journals urinary malignancy Scopus journals urinary malignancy PubMed journals urinary malignancy medical journals urinary malignancy free journals urinary malignancy best journals urinary malignancy top journals urinary malignancy free medical journals urinary malignancy famous journals urinary malignancy Google Scholar indexed journals diagnostics articles diagnostics Research articles diagnostics review articles diagnostics PubMed articles diagnostics PubMed Central articles diagnostics 2023 articles diagnostics 2024 articles diagnostics Scopus articles diagnostics impact factor journals diagnostics Scopus journals diagnostics PubMed journals diagnostics medical journals diagnostics free journals diagnostics best journals diagnostics top journals diagnostics free medical journals diagnostics famous journals diagnostics Google Scholar indexed journals Cancer articles Cancer Research articles Cancer review articles Cancer PubMed articles Cancer PubMed Central articles Cancer 2023 articles Cancer 2024 articles Cancer Scopus articles Cancer impact factor journals Cancer Scopus journals Cancer PubMed journals Cancer medical journals Cancer free journals Cancer best journals Cancer top journals Cancer free medical journals Cancer famous journals Cancer Google Scholar indexed journals microenvironment articles microenvironment Research articles microenvironment review articles microenvironment PubMed articles microenvironment PubMed Central articles microenvironment 2023 articles microenvironment 2024 articles microenvironment Scopus articles microenvironment impact factor journals microenvironment Scopus journals microenvironment PubMed journals microenvironment medical journals microenvironment free journals microenvironment best journals microenvironment top journals microenvironment free medical journals microenvironment famous journals microenvironment Google Scholar indexed journals tumors articles tumors Research articles tumors review articles tumors PubMed articles tumors PubMed Central articles tumors 2023 articles tumors 2024 articles tumors Scopus articles tumors impact factor journals tumors Scopus journals tumors PubMed journals tumors medical journals tumors free journals tumors best journals tumors top journals tumors free medical journals tumors famous journals tumors Google Scholar indexed journals
Article Details
1. Introduction
Urothelial bladder carcinoma (UBC) is a common urinary malignancy [1]. It represents more than 10% of new cancer diagnostics, worldwide [2]. It is often found in elderly males (sex-ratio males to females of 3.5:1), with a median age at diagnostic of 65 years [3]. Environmental factors (like air pollution and tobacco use) and genetics are considered responsible for the significant morbidity and mortality associated with this disease [4]. UBC originates in the epithelial layer of urothelium, a highly specialized, multi-stratified tissue existing in the distant urinary tract [5]. The normal urothelial tissue characteristics may explain the numerous forms and variants of UBC and the complexity of the histologic diagnostic [6]. A precise histologic characterization (types, forms and variants) of any UBC is important as it will ultimately dictate the patient prognostic under treatment. The dominant UBC histologic type (90%) is transitional carcinoma [7]. More than 70% of transitional carcinomas are low grade, with a uniform papillary architecture and are Non-Muscle Invasive (NMIBC), but are still susceptible to recurrence and progression [8]. Muscle-invasive BC (MIBC) have a marked tendency to histological diversity and is frequently associated with stromal and inflammatory reaction, fibroblastic proliferation and fibrosis [9], all factors that will add supplementary challenges to the routine diagnostic. MIBC demonstrates an aggressive clinical behavior even in early stages [10]. Improving UBC grading precision (both clinical and pathological) was a continuous and strenuous exercise over time with results that are still under evaluation [11-14]. The advent of new, specific, histologic biomarkers provided more clarity in pathology grading [15] but both immunohistologic and biomolecular procedures are costly, time consuming and sometimes subjective, most of these methods having a high degree of inter-observational variability [16,17]. In early MICB, pathological grading remains the mainstay for treatment option but a precise differentiation between early forms is not always easy as changes at muscularis propria level are often subtle [18]. Optical detection of muscularis propria invasion in early MIBC can be difficult to substantiate as it can be subject of multiple technical challenges but the right diagnostic will dictate an adequate therapeutic approach and will show tremendous prognostic value. The capacity to interact with the surrounding microenvironment is a critical characteristic of any solid malign tumor [19]. The tumoral microenvironment (TME) is a complex and continuously evolving tumor entity that influences tumoral invasiveness and progression [20]. Components of tumor microenvironment can be recognized within the routine microscopic exam but a precise visual characterization of the tumor based on these characteristics is difficult as is subjective, with serious inter and intra observational variability. One important actor in the complex TME landscape is tumoral surrounding stroma, a protective canvas that increase tumor aggressiveness and shield the tumor against treatment [21]. It is generally accepted [22] that stromal development (enhanced local vasculature, modified cellularity, increased inflammatory response, and imbalanced protease activity) often precedes tumor progression. It was also signaled that, at least for some tumors, an active stroma may play a protective role against cancer progression [23, 24]. The ratio between tumor cells and stroma (tumor stoma ratio - TSR) can be measured. A low ratio (more stroma, less tumor) characterized most of the aggressive tumors. The ratio was validated in many solid tumors as an independent prognostic predictive factor [25]. An accepted definition of a high TSR is when more than 50% of tumor are represented by cells. In many tumors, a high TSR was associated with a better clinical prognosis.
Compared to other solid cancers (colorectal, hepatopancreatic, breast), the TSR concept was not extensively tested yet in UBC [26]. Previous published research suggested that a low TSR (a high stroma presence) may reflect a poor prognostic over time [27]. There are several reasons for adopting TSR as part of the routine diagnostic toolkit in UBC [28]. It is measured in Hematoxylin-Eosin (HE) stained slides. It had a demonstrated prognostic predictive value in many solid tumors. As no other stains are needed, there is a possibility to better control diagnostic costs. In early MICB, TSR may support the histologic diagnostic and provide objective arguments for making the differentiation between pT2A and pT2B, simple. The challenge comes from visual quantification of TSR, a method that is subjective and time consuming even for highly specialised uro-pathologists. A machine learning analysis (using a technique named semantic segmentation) can differentiate between tumor and stroma and provide an objective ratio, fast and at low costs. Our research interest is to evaluate how precision pathology tools (machine learning segmentation of whole slide images - WSI) may improve quantification of the tumor-stroma ratio in early muscle invasive bladder tumors and may increase histologic diagnostic prognostic value.
2. Materials and Methods
Data selection. 10 cases with pathological stage (p)T2A UBC diagnostic were selected from The Cancer Genome Atlas (TCGA), a public, open database available at National Institute of Health-National Cancer Institute (access @ https://portal.gdc.cancer.gov). Each case had at least one diagnostic WSI and an associated pathology report that served as “ground truth” (pathology data available @ https://cancer.digitalslideatlas.org). The ten selected cases were carefully matched (age, sex, smoking status) with 10 (p)T2B cases, from the same databases. The (p)T stage (based on AJCC criteria) was decided based on the degree of muscularis propria invasion (inner or outer half). All sampled cases were classified, based on cellular architecture, as high grade. Epidemiologic, clinical and genomic biomarkers information was available for all selected cases. Cases with an extreme number of genetic mutations (both at high and low ends) were excluded.
Data processing 20 WSI were downloaded and evaluated using QuPath software [29]. At a 10x magnification (1 µm=1 pixel), a 1024/1024 pixels rectangular region of interest (ROI) was selected by both investigators, in consensus. Selected ROIs images (area=1048576 pixels2) were deconvoluted (normalized) using the FIJI software [30]. Only the hematoxylin channel image was used for semantic segmentation (figure 1), under 3 labels (tumor, stroma, other). For segmentation we used a dedicated WEKA machine learning platform plugin [31]. The area covered under each label was finally measured and recorded using FIJI capacities. In order to avoid segmentation overfitting, the classifier was initially trained on a single pT1 image and then tested on another single pT3 image (pT3) (procedure known as the data veracity test). Once the classifier was considered performant, it was used for the segmentation of all of the 20 selected cases, without any other alteration. TSR was calculated using only tumor and stroma areas, without considering the “other” area label. All cases were blindly evaluated by both authors (visual WSI exam) looking for tumoral stromal area approximation (less or more than 50% of tumoral cellularity). A second objective of the human exam was to confirm the initial diagnostic criterion (invasion of muscularis propria).
Ethics Data used in this study was coming from an open, public database where cases are totally anonymized. The de-identification process was performed by data curators. Our study followed the Canadian ethics research provisions for secondary data use studies (TCPs 2(2018)) and the principles specified in Declaration of Helsinki. The overall TSI research was evaluated and approved by the IRB of “Sfântul Ioan” Hospital Bucharest (28827/Nov 2021).
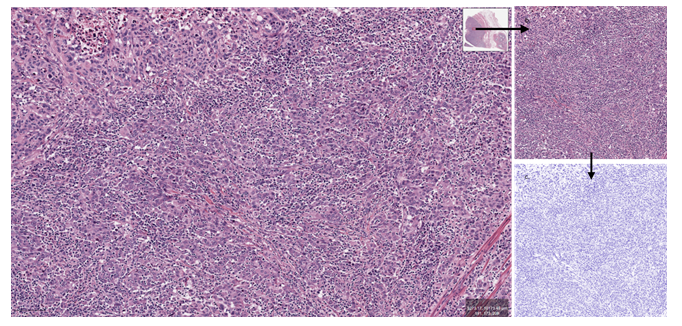
Figure 1: Image preparation (HE). a. Selecting a ROI from the initial WSI image (QuPath) (bar=100µm). b. Sampled ROI (1024x1024px, 1pixel=1µm). c. Normalized (deconvoluted) image (FIJI).
3. Results
10 pT2A cases (80% males, 40% smokers, mean age 68.5±8.43 years) were carefully matched with 10 pT2B cases (80% males, 40% smokers, mean age 70.7±8.7 years) from TCGA-BLCA project (412 cases). From each WSI, a region of interest (ROI) was selected, in consensus. The ROI image was sampled and prepared. Finally, images were segmented using a previously trained segmentation plugin and areas were measured. The pT2A group had a mean tumor area of 58.25%, a mean stroma area of 38.6% and a 3% area for “other”. pT2B group had a mean tumor area of 40.8% (“tumor”), 57% (“stroma”) and 2.1% (“other”) on similar ROIs (table 1, figure 2). All cases were papillary, high grade UBC and all were considered histological grade 2. In pT2A group, the multi-tumor diagnostic was an exception. In pT2B, 2 cases were squamous (>10%) and 3 cases had a CIS associated diagnostic. In terms of invasiveness, in pT2A group, 1 case was multi-centric. In pT2B 5 cases were characterized as multicentric, 3 cases demonstrated vascular and 1 lymphatic spread.
Avoiding overfitting is an important step in using machine learning segmentation. In our case, the classifier was trained separately on one pT1 case (male, 50 years, non-smoker, low grade tumor). It showed 63% tumoral area and a stromal area of 27%. Then the classifier was tested on a pT3 case (male, 57 years, non-smoker, high grade tumor) with a measured tumoral area of 35% and a stromal area of 64% (similar ROI). Both cases used for classifier training/testing were with squamous traits (>10%). A blinded visual exam of WSI established (based on the 50% tumoral limit) that results were concordant with machine learning measurements only in 40% of pT2A cases and in 50% of pT2B cases, with a high observational test-retest variability (50-55%). 9/10 cases in pT2A and only 5/10 cases in pT2B cohort were alive at 3 years after the primary MIBC diagnostic (figure 3).
|
Cohort pT2A (10 cases) |
Cohort pT2B (10 cases) |
One pT1 case for training |
One pT3 case for testing |
|
|
Diagnostic |
T2A |
T2B |
T1 |
T3 |
|
Sex |
80%M |
80%M |
M |
M |
|
Age |
68.5±8.43 |
70.7±8.7 |
50 |
57 |
|
Smoker |
40% |
40% |
No |
No |
|
Histo |
100% High |
100% High |
Low |
High |
|
Infiltration MM |
Inner |
Outer |
No |
Yes |
|
Associated Histology |
1 nodular |
2 squamous, 3 CIS |
Squamous |
Squamous |
|
Spread |
1 multicentric |
5 multicentric, 3 vascular, 1 lymphatic |
No |
Multicentric |
|
Visual approximation of ROI (blinded) |
40%+ tumor |
50%+ tumor |
55%+ tumor |
50%+ tumor |
|
Tumor area |
58.25%* |
40.8%* |
63% |
35% |
|
Stroma Area |
38.6%* |
57.1%* |
27% |
64% |
|
Other |
3.15%* |
2.1%* |
10% |
1% |
|
Survival |
9 Alive / 1 Death |
5 Alive / 5 Death |
Alive |
Deceased |
|
TSR |
1.48±0.21# |
0.72±0.11# |
2.3 |
0.54 |
|
* Averaged data, 10 cases, MM=muscularis propria, CIS = Carcinoma in situ, #p<0.0001. Visual approximation used a five points Visual Analog Scale |
||||
Table 1: TSR, areas measurements and demographic data in pT2A / pT2B cohorts the classifier was trained in one pT1 and tested in another pT3 case.
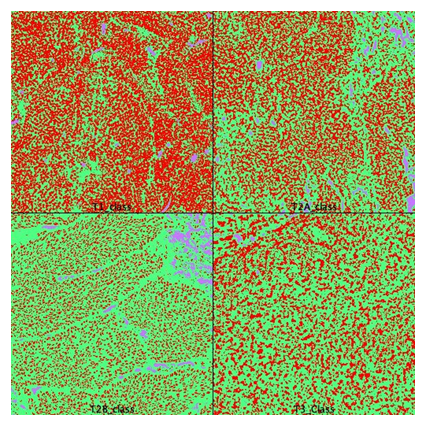
Figure 2: Area calculation on segmented images. pT1 was used for classifier training. pT3 was used for classifier validation. T2A and T2B areas were quantified and averaged. Red=tumor, green=stroma, blue=other (FIJI, WEKA segmentation plugin).
3. Discussion
UBC is a common tumor, frequently encountered in elderly males. The tumor may show a large diversity of histologic forms and variants that will dictate invasiveness and progression. A precise histologic diagnostic is required as histology remains the main pillar of treatment decision. Finding new biomarkers that will better characterize UBC in an objective, reproductible way, is a constant research effort. TME gained researchers’ interest as it demonstrated to be an effective way to characterize solid tumors. TSR, a histologic marker of TME, proved to be a reliable indicator of tumor aggressiveness, an independent prognostic indicator in many solid tumors. In most of the solid tumors (lung, breast, colorectal) distinction between low and high stroma can be done by visual approximation using simple visual microscopy tools but is still imprecise and time consuming. In UBC, the limit between high and low TSR is difficult to approximate using visual measurements (cut-off limit is 50% - Micke op cit. [29]). Our research objective was to evaluate how precise pathology (machine learning seg-mentation of WSI) [32] will improve TSR quantification in MIBC. We sampled 10 UBC pT2A cases from TCGA-BLCA project at NHI-NCI. Cases have at least one diagnostic WSI. Every selected case was carefully matched (sex, age, smoking status) with a pT2B case from the same database. All WSI images were obtained from tissues that were processed following a unique, highly standardized technique and have associated a complete pathology report that served as ground truth for diagnostic. As genetic data was also available, cases with a particular high or low load of genetic mutations were discarded from initial selection. All cases were high grade papillary transitional carcinomas. Pathologic pT2 stage was defined based on muscularis propria invasion (inner vs outer half) and was well documented in the existing pathology report. All tumors were considered grade 2. All images were examined at 10x (1pixel=1 µm). From a tumor area that showed a high tumor and a low artifact load, a ROI of 1024x1024 pixels was selected by both authors, in consensus. ROI images were normalized (deconvoluted) and then segmented using a pre-trained classifier, under three labels (tumor, stroma, other). In order to avoid over-fitting, the classifier was initially trained on a pT1 case and then tested on a pT3 case.
The classifier was used in all 20 selected cases without any other alteration. After segmentation, each area was measured. The calculated mean TSR in pT2A group was significantly different (p<0.0001) compared to T2B cohort mean. Repeated tests showed no results variability. We performed an individual blinded visual diagnostic on downloaded WSI. Tumor/stroma area approximation using a visual analogue scale (%, from low to high) showed a low precision (40-50%) and a high intra and inter-observational diagnostic variability (50%-55%). The same subjectivity of visual analogue scale in difficult to diagnostic UBC was confirmed by other published data [33]. The survival analysis of selected subjects confirmed the clinical predictive value of TSR marker in early MIBC. 90% of T2A subjects were alive at 3 years after diagnostic (1 dead male, 74 years old). In T2B, only 40% of cases were still alive at 3 years from diagnostic (4 males, mean age at diagnostic 69 years) (Figure 3).
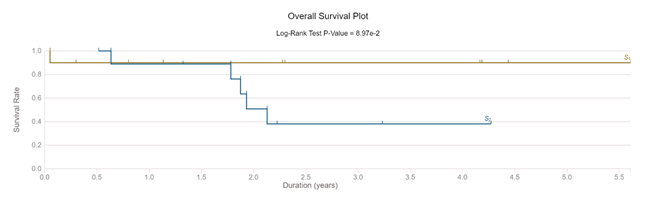
Figure 3: Survival of selected cases (pT2A compared with pT2B)
From a genomic perspective, the pT2A group showed a rather moderate altered genomic profile with variations in 2 genes that affected more than 50% of cases (MUC16 50%, TP53 60%). For the pT2B cohort, the genomic picture was very rich, subjects showing multiple genes mutations: TTN (90%), TP53 (80%), MUC16 (60%), DNAH5 (60%), HMCN1 (50%), FBN2 (50%) and RYR2 (50%). TTN and TP53 are genes that affected most of the subjects included in the TCGA-BLCA dataset (>50% of all subjects). TP53 is a recognized prognostic indicator in bladder cancers. As all of cases were high grade, invasive transitional carcinomas, a low number of cases showed a FGFR3 mutation (30% in pT2A and none in pT2B cohort) [34]. The low number of cases used in our study did not allow any prediction concerning the effects of specific gene mutations on survival.
Machine learning TSR quantification was fast, precise and objective. There was no difference in results when similar ROIs were tested with the same classifier. In contemporary clinical practice, UBC is characterized using immunohistochemical diagnostics that are expensive, time consuming and not very precise as quantification is performed using a visual analogic scale (fast score) [35]. Routine use of machine learning techniques on immunohistochemical stained slides is controversial as the biomarker quantification is considered not iso-stoichiometric [36]. Quantification STR on HE slides can also be automated, fact that can reduce diagnostic time and costs. The main limitation of our study is the low number of cases evaluated (proof of concept). TCGA-BLCA is structured as a genomic dataset with rich attached pathology information (412 UBC cases). The dataset is built mainly on advanced carcinomas (60% of cases being pT3 or more) and early cases are rare. We considered that the quality of data (strict inclusion criteria, well standardized histologic preparing techniques and the existence of a ground truth – a pathology report) are benefits that increased the validity of machine learning quantification, our main study objective. The complex demographic and genomic associated info were also seen as advantages for our study.
5. Conclusions
A precise machine-learning measurement of TSR in MIBC is possible. When performed in highly standardized WSI, the difference between pathology stage 2A and 2B ratios was highly significant. TSR reflected well the tumor aggressiveness and had a true prognostic prediction value. As the number of cases was low, any association between tumor staging/grading and genomic picture was not possible but it we have seen a clear difference between the genomic picture of the two analyzed cohorts. Larger studies focusing on real world pathologic data are needed for method validation and for any possible wide genomic correlations.
Author Contributions
Both authors participated equally in data research, data interpretation, article drafting and review.
Funding:
This research received no external funding
Institutional Review Board Statement
The whole TSR histologic investigation was approved by “Sfântul Ioan” Hospital, Bucharest Romania, approval 28827/Nov 2021.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest
Data Availability Statement
All data used in this study was coming from public domain and is available for consultation upon request.
Acknowledgments
Both authors want to thank TCGA-BLCA dataset curators for the high-quality data provided (National Institute of Health @ https://gdc.cancer.gov/publication-tag/tcga-blca).
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol 71 (2016): 96-108.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 66 (2016): 7-30.
- Ferlay J, Bray F, et al. Globocan 2000: cancer incidence, mortality and prevalence worldwide, IARC Press, Lyon, (2001).
- Yuen-Chun JT, Huang J. Global Trends of Bladder Cancer Incidence and Mortal-ity, and Their Associations with Tobacco Use and Gross Domestic Product Per Capita, European Urology 78 (2020): 6.
- Kamat AM, Hahn NM, Jason AE, et al. Bladder cancer. Lancet 388 (2016): 2796-2810.
- Khandelwal P, Abraham SN, Gerard A, et al. Cell biology and physiology of the uroepithe-lium. Am. J. Physiol. Renal. Physiol 297 (2009): F1477-F1501.
- DeGeorge KC, Holt HR, Hodges SC. Bladder Cancer: Diagnosis and Treatment. Am Fam Physician 96 (2017): 507-514.
- Abdollah F, Gandaglia G, Thuret R, et al. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: A trend analysis. Cancer Epidemiol 37 (2013): 219-225.
- Hemamali S, Peta F, David P, et al. Significance of Stromal Reaction Patterns in Invasive Urothelial Carcinoma, American Journal of Clinical Pathology 123 (2005): 851-857.
- Amin MB. Histologic variants of urothelial carcinoma: diagnostic, therapeutic and prognostic implications. Modem Pathol 22 (2009): S96-118.
- Manini C, Lopez JI. Unusual Faces of Bladder Cancer, Cancers 12 (2020): 3706.
- Epstein JI, Amin MB, Reuter VR, et al. The World Health Organization/International Society of Urological Pathology consensus classification of urothelial (transitional cell) neoplasms of the urinary bladder. The American Journal of Surgical Pathology 22 (1998): 1435-1448.
- Fletcher CDM, Unni K, Mertens F. World Health Organization classification of tumours. Pathology and genetics of tumours of soft tissue and bone. IARC press (2004).
- Moch H, Cubilla Al, Peter AH, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. European urology 70 (2016): 93-105.
- van_der_Kwast T, Liedberg F, et al, International Society of Urological Pathology Expert Opinion on Grading of Urothelial Carcinoma, European Urology Focus (2021).
- Kobayashi T, Owczarek TB, McKiernan JM, et al. Modelling bladder cancer in mice: opportunities and challenges. Nat Rev Cancer 15 (2015): 42-54
- Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumour markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology 61 (2003): 109-118.
- Fathollah Keshvar M, Davis CJ Jr, Sesterhenn IA. Histological typing of urinary bladder tumours.
- Anderson NM, Simon CM. Tumor microenvironment Curr Biol 30(2020): R921-R925.
- Yuan Y, Jiang Y-C, Sun C-K, et al. Role of the tumor microenvironment in tumor progression and the clinical applications (Review). Oncol Rep 35 (2016): 2499-2515.
- Valkenburg KC, de Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol 15 (2018): 366-381.
- Micke P, Tman A, Tumour-stroma interaction: cancer-associated fibroblasts as novel targets in anti-cancer therapy?, Lung Cancer 45 (2004).
- Panayiotou H, Orsi NM, Helen HT, et al. The prognostic significance of tumour-stroma ratio in endometrial carcinoma. BMC Cancer 15 (2015): 955.
- Bever KM, Sugar EA, Elain B, et al. The prognostic value of stroma in pancreatic cancer in patients receiving adjuvant therapy. HPB (Oxford) 17(2015): 292-298.
- van Pelt GW, Kjær-Frifeldt S, Mesker WE, et al., Scoring the tumor-stroma ratio in colon cancer: procedure and recommendations. Virchows Arch 473 (2018): 405-412.
- Vrabie CD, Gangal M. Precise quantification of Tumor-Stroma Index in Bladder Cancer - a 12 months scoping review (2022).
- Micke P, Strell C, et al The prognostic impact of the tumour stroma fraction: A machine learning-based analysis in 16 human solid tumour types. EBioMedicine 65 (2021):103269.
- Smit M, van Pelt G, Annet R, et al. Uniform Noting for International Application of the Tumor-Stroma Ratio as an Easy Diagnostic Tool: Protocol for a Multicenter Prospective Cohort Study. JMIR Res Protoc 8 (2019): e13464.
- Bankhead P, Jose F, Yvonne D, et al. QuPath: Open-source software for digital pathology image analysis. Scientific Reports (2017).
- Schindelin, J, Arganda-Carreras I, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9 (2012): 676-682.
- Arganda-Carreras I, Kaynig V, Curtis R, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33 (2017): 2424-
- Djuric U, Zadeh G. Precision histology: how deep learning is poised to re-vitalize histomorphology for personalized cancer care, Precision Oncology (2017) 1:22.
- Pellucchi F, Freschi M. IbrAHIM B, et al. Clinical Reliability of the 2004 WHO Histological Classi-fication System Compared With the 1973 WHO System for Ta Primary Bladder Tumors. The Journal of Urology 186 (2011): 2194-2199.
- Guancial EA, Werner L, Bellmunt J, et al. FGFR3 expression in primary and metastatic urothelial carcinoma of the bladder. Cancer Med 3 (2014): 835-844.
- Harvey JM, Clark GM, Osborne CK, et al. Estrogen Receptor Status by Immunohistochemistry Is Superior to the Ligand-Binding Assay for Predicting Response to Adjuvant Endocrine Therapy in Breast Cancer. J Clinical Oncology 17 (1999): 1474-1481.
- Landini G, Martinelli G, Piccinini F. Colour deconvolution: stain unmixing in histological imaging, Bioinformatics 37 (2021): 1485-
