Identification of Itch Mediators and Its Novel Pathway: Implications for Understanding Radiation-Induced Skin Effects
Article Information
Yuan-Hao Lee1, Youping Sun2*
1Department of Radiology, Wan Fang Hospital, Taipei Medical University, Taipei 116, Taiwan
2Center for Radiological Research, Department of Radiation Oncology, Columbia University Medical Center, New York, NY 10032, USA
*Corresponding Author: Youping Sun, Center for Radiological Research, Department of Radiation Oncology, Columbia University Medical Center, New York, NY 10032, USA, Tel: +1 617-9354894.
Received: 14 October 2016; Accepted: 19 October 2016; Published: 24 October 2016
View / Download Pdf Share at FacebookAbstract
Background: Radiation-induced skin reactions often lead to itching and pain. While many cytokines bind and activate nociceptors to initiate pain and the precedent itch upon radiation-induced skin desquamation, little is known that radiotherapy-induced itch can arise from the activation of Notch signaling. With regard to the enhanced ransactivation activity of Notch after irradiation and the Notch-induced intense scratching of mice, we anticipated that a downstream effector of Notch mediates the itch sensation.
Aim: The purpose of this study was to investigate the role of Notch receptor in mediating radiation-induced itch. Methods: Primary human keratinocytes, HEK293T/17, and A549 cells with or without NICD (Notch intracellular domain) overexpression were applied for analytical assays, including qRT-PCR, SDS-PAGE, immunoprecipitation, and luciferase assays, for delineating the relationship between Notch and Cathepsin Sas well as the effect of Cathepsin S on the itch-inducing stimulus, thymic stromal lymphopoietin (TSLP).
Results: Cathepsin S mRNA and protein were upregulated in response to the overexpression of Notch intracellular domains in primary human keratinocytes. A consensus binding site for Notch-regulated transcription factor CBF-1 (C-repeat/dehydration-responsive element binding factor 1) was identified in the promoter region of Cathepsin S. Along with the observation that Cathepsin S immunoprecipitated with PAR2 (protease-activated receptor 2), TSLP Arch Clin Biomed Res 2016; 1 (1): 9-19 10 was found dramatically increased with Cathepsin S in primary human keratinocytes.
Conclusions: This study validates that Cathepsin S is a downstream target gene of Notch and mediates itch by upregulating TSLP. The found mechanism may be targeted for improving patients’ quality of life during radiotherapy.
Keywords
Notch signaling; Cathepsin S; Itch sensation
Article Details
1. Introduction
Radiation-induced skin reactions experienced by patients treated with radiotherapy often incur sequential radiation effects, i.e. itch, pain, delays in treatment could subsequently decrease in patients’ life quality [1]. While many cytokines bind and activate nocireceptors as well as drive the degeneration of the nervous system to initiate pain upon radiation-induced skin desquamation, a sensation of precedently accompanied itch are usually suppressed by scratches and antagonized by pain sensation. Preventive methods of radiation-induced skin reactions, however, have not yet completely diminished itch sensation.
In response to radiation, a multitude of signaling pathways in and between cells are either activated or suppressed for cell survival or the best cell-fate choices. Among all regulated pathways, Notch signaling is often activated to confer cellular radioresistance [2,3]. A series of relevant studies also reveal that impaired Notch ubiquitination and lysosomal degradation can lead to intense scratching and an autoimmune disease in mice [4-7]. In this respect, we investigated the role of the Notch receptor in mediating itch sensation.
Cathepsin S, a cysteine protease, appears to involve in radioresistance and cause itching [8,9]. As a member of the papain family, Cathepsin S is important for intracellular and extracellular degradation and the presentation of antigens on immune cells [10,11]. Kempkes C et al. has shown that Cathepsin S evokes itch through the activation of PAR2 (protease-activated receptor 2) in human skin [12]. In mice, researchers have shown that Cathepsin S activates MrgprC11, a member of Mas-related G-protein-coupled receptors, and evokes receptor-dependent scratching [9]. Nevertheless, it is unknown whether Notch-induced itching can be mediated by Cathepsin S.
There are four Notch receptor paralogs, i.e., Notch 1, 2, 3 and 4, and all the four paralogs have been associated with radioresistance under different pathological circumstances [13-15]. In the normal physiological condition, Notch receptors as transmembrane receptors are activated upon enzymatic cleavage and nuclear translocation of the Notch intracellular domain (NICD) to control a broad spectrum of cell-fate decision [3,16]. The NICD carries nuclear localization signals, which enable it to directly participate in a transcriptional complex and drive Notch-dependent transcription [16]. Here we describe a novel mechanism by which Notch activation induces itching through the upregulation of Cathepsin S and an epithelial-derived cytokine, TSLP (thymic stromal lymphopoietin).
2. Methods
2.1 Cell Culture
Primary human keratinocyte epithelial cells were purchased from Lonza (Cat # 192627, Lonza; Basel, Switzerland) and maintained in Defined Keratinocyte-SFM (Cat # 10744, Invitrogen; Carlsbad, CA, USA). HEK293T/17 cells were purchased from the ATCC (ATCC® CRL-11268, Manassas, VA, USA). These cells were grown in DMEM medium (Cat # 21063-029, Invitrogen; Carlsbad, CA, USA), supplemented with 10% fetal bovine serum.
2.2 Plasmids
HA?tagged human NICD1 (Notch1 intracellular domain) and human NICD3 (Notch3 intracellular domain) were kindly provided by Dr. Lizi Wu [19]. Ha-tagged mouse NICD4 expression vectors were made through PCR Pfu synthesis by inserting Notch4 ICD (intracellular domain) sequences into pcDNA3 vector (Invitrogen; Carlsbad, CA, USA). The cloning of NICD4 (Notch4 intracellular domain) with pcDNA 3.1 pEF1/V5His TOPO TA expression vector was previously made and validated [20].
2.3 Luciferase Reporter Assay
Human Cathepsin S promoters with different length (a full 2.8-kb fragment with a CBF binding site, a 2.6-kb or a 1-kb fragments without the CBF binding site) were cloned into the vector pGL-4-14 luciferase (Promega; Madison, WI, USA) using XhoI and HindIII restriction sites. The human genetic DNA was purchased from life technology as a template to clone Cathepsin S promoter region. Human Cathepsin S promoter was generated through PCR using Pfu polymerase (Stratagene; La Jolla, CA, USA) and cloned into the luciferase expression vector pGL4-14. The forward primer sequence, 5’-GCGCTCGAGGGAGGCTGAGG TGGGCAGATTACCTATGGT-3’, was used to generate an XhoI cutting site. The reverse primer sequence, 5’-GCGAAGCTT-GCGGGATCCTTCAAAGCAACAAGGAG-3’ was applied to create a HindIII site. pGL4-14 was cut by both XhoI and HindIII while the ligations were performed as described previously [20]. The 2.6-KB Cathepsin S-luciferase construct was amplified by a forward primer with the sequence of 5’-GCGCTCGAGA AATAAAATAAGATTGGCCATGTACTGATCATTGTTGAAAC-3’, which gave rise to an EcoRI cutting site along with the above mentioned reverse primer. The 1.0-KB Cathepsin S-luciferase construct was amplified with a forward primer in the sequence of 5’-GGGGGTACCCCCCACACCT CCCACAAATCACTAAGAAAATGCAACAATAC-3’, which generated an XhoI cutting site along with the above mentioned reverse primer. Luciferase assays were performed according to the manufacturer’s (Promega; Madison, WI, USA) protocol as previously described [20].
2.4 Protein Extraction and Antibodies
Cells were lysed as previously described [16]. Fifty micrograms of protein was subjected to SDS-polyacrylamide gel electrophoresis (Invitrogen; Carlsbad, CA, USA) as previously described [21]. Applied antibodies included anti-FLAG-HRP (Cat # A8592, Sigma; St. Louis, MO, USA), anti-Notch4 (Cat # 07-189, Millipore; Bedford, MA, USA), anti-HA (Cat # 11867423001, Roche; Basel Switzerland), anti-Cathepsin S (Cat # GTX114350, GeneTex; Irvine, CA, USA).
2.5 Transfection
Primary human keratinocyte epithelial and A549 cells (in the density of 106 cells on a 60-mm-diameter plate) were
transfected with 2?g HA-NICD1, HA-NICD2, or HA-NICD4 expression vector, individually with or without using Lipofectamine 2000 reagent (Invitrogen; Carlsbad, CA, USA) according to the manufacturer’s instruction.
2.6 Co-Immunoprecipitation of PAR-2 with Cathepsin S
HEK 293T/17 cells were transfected with plasmids encoding Flag-tagged PAR2 and/or Cathepsin S expression vector(s). After forty-eight hours of transfection, cells were treated with 30?M MG-132 for 5 hours. Co-Immunoprecipitations were performed on cell lysates with an anti-FLAG antibody, and protein interactions were monitored by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) as described in the following section.
2.7 SDS-PAGE
Cells were harvested in RIPA buffer (150mM NaCl, 10mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 1% deoxycholate, 5mM EDTA) supplemented with the protease inhibitor cocktail set 1 (Calbiochem). The total protein concentration was determined with BCA Protein Assay Kit (Pierce Biotechnology Inc.; Waltham, MA, USA). Protein pellets (50 ?g each time) were loaded on 4-20% gradient SDS-PAGE gels and transferred to nitrocellulose membranes using the iBlot Transfer System (Invitrogen; Carlsbad, CA, USA). After membrane blocking with blocking buffer (5% non-fat milk in TBS with 0.1% Tween20 (TBS/T) for one hour at room temperature, primary antibodies were applied for incubation. Followed by membrane washing with TBS/T and incubation with the secondary antibody conjugated to HRP, the targeted protein bands were visualized using the ECL Western Blotting Detection System (Cat #: RPN2132, GE Healthcare; Pittsburgh, PA, USA).
2.8 RNA Isolation, cDNA Synthesis and Real Time qRT-PCR
The method of RNA isolation, cDNA synthesis and RT-PCR were conducted as previously described [20,21]. Briefly, total cellular RNAs were obtained from cell lines using TRIzol (Invitrogen; Carlsbad, CA, USA). cDNAs were synthesized from 1 ?g total RNA with random primers and a SuperScript II kit (Invitrogen; Carlsbad, CA, USA) according to the manufacturer’s protocol. RT reactions were performed with a SuperScript II RT kit (Invitrogen; Carlsbad, CA, USA). The positive control, HPRT (hypoxanthine-guanine phosphoribosyltransferase) transcript, was synthesized with the following paired primers. The forward and reverse primer sequences were 5’-GACACTGGCAAAACAATGCAGAC-3’ and 5’-CAGTTTCACTAATGACACATTCATG-3’, respectively. The forward and reverse primer sequences of Cathepsin S (gene accession number: P25774) were 5’-CATGAACCACTGGGAGACATG-3’ and 5’-ACACCAGCTTTCCTG TTTTCAGC-3’, respectively.
2.9 Statistical Analysis
Quantitative data were represented as the mean value of at least three independent experiments. Statistically
significant differences between groups were determined by the Student-t test. Analytical results of p-values less than 0.05 were considered statistically significant and indicated in the figures with asterisks (*).
3. Results
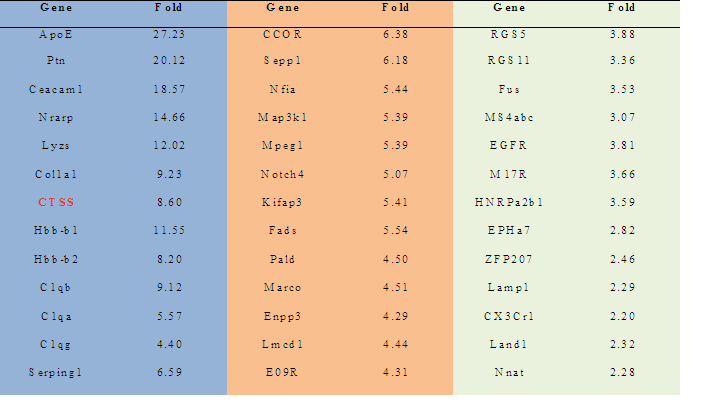
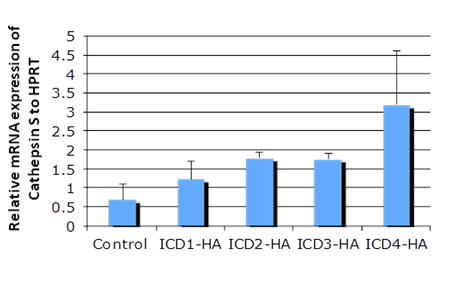
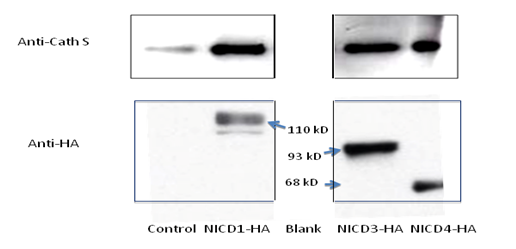
3.1 Notch 4 Activation Upregulated Cathepsin STo determine whether NICD1, 3 and 4 can upregulate Cathepsin S expression, primary keratinocytes were transfected with individual HA-tagged Notch intracellular domains and were characterized by RT-PCR and SDS-PAGE assays (Figure 1 A & B). NICD1, 3 and 4 increased both the mRNA and protein levels of Cathespin S to different extents, compared with those of the control (Figures 1A and 1B). Correspondingly, an 8.6-fold upregulation of Cathepsin S was found in mammary tumors expressing MMTV (mouse mammary tumor virus)-NICD4 (Table 1) compared with that found in large T -/- & BRCA1-/- mammary tumors. These results indicate that Cathepsin S can be upregulated by Notch activation, particularly Notch4 activation.
3.2 Cathepsin S is a Direct Transcriptional Target of Notch
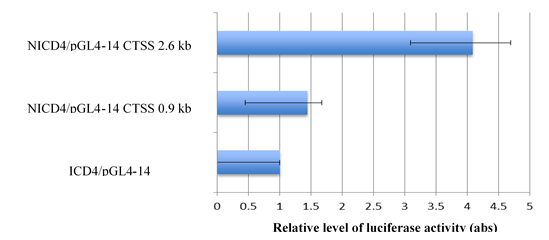
To find out whether NICD4 activates Cathepsin S, A549 cells were transfected with plasmids expressing NICD4
and/or luciferase under a human Cathepsin S promoter in a reporter assay, respectively. By measuring the luciferase activity of cells driven by different regions of Cathepsin S promoter, we observed different levels of NICD4-mediated transactivation at different lengths of Cathepsin S promoter (Figure 2). Compared with the control group, cells transfected with the 0.9-kb upstream region of the Cathepsin S promoter expressed about 1.4 fold of luciferase. The expression was four times more than that of the control group when cells were transfected with a 2.6-kb long promoter, which harbored a CBF-binding site. This result indicates that Cathepsin S is a direct transcriptional target of NICD4, which may bind to the specific CBF-1 region of Cathepsin S promoter.
Table 1: The ratio of cDNA expression : MMTV-NICD4 & Wap-NICD4 expressing Tumors vs. Large T -/- & BRCA1-/- Mammary Tumors
3.2 Cathepsin S is a Direct Transcriptional Target of Notch
To find out whether NICD4 activates Cathepsin S, A549 cells were transfected with plasmids expressing NICD4
and/or luciferase under a human Cathepsin S promoter in a reporter assay, respectively. By measuring the luciferase activity of cells driven by different regions of Cathepsin S promoter, we observed different levels of NICD4-mediated transactivation at different lengths of Cathepsin S promoter (Figure 2). Compared with the control group, cells transfected with the 0.9-kb upstream region of the Cathepsin S promoter expressed about 1.4 fold of luciferase. The expression was four times more than that of the control group when cells were transfected with a 2.6-kb long promoter, which harbored a CBF-binding site. This result indicates that Cathepsin S is a direct transcriptional target of NICD4, which may bind to the specific CBF-1 region of Cathepsin S promoter.
Figure 1a: Cathepsin S is a target gene of Notch. The mRNA level of individual Notch intracellular domain (NICD) was overexpressed in primary keratinocytes (panel A); the protein levels of Cathepsin S were increased with NICD overexpression (panel B). Abbreviations NICD1: Intracellular domain of Notch1, NICD3: Intracellular domain of Notch3, NICD4: Intracellular domain of Notch4, Cath S: Cathepsin S
Figure 1b: Cathepsin S is a target gene of Notch. The mRNA level of individual Notch intracellular domain (NICD) was overexpressed in primary keratinocytes (panel A); the protein levels of Cathepsin S were increased with NICD overexpression (panel B). Abbreviations NICD1: Intracellular domain of Notch1, NICD3: Intracellular domain of Notch3, NICD4: Intracellular domain of Notch4, Cath S: Cathepsin S
Figure 2: Cathepsin S promoter has a CBF binding site and can be activated by NICD4. Cathepsin S promoter at different lengths (2.6 kb and 0.9 kb) was transactivated by NICD4 and increased luciferase expression in A549 cells. The 2.6-kb promoter harbored a CBF1-binding site, which may be bound by NICD4. Abbreviation NICD4: Intracellular domain of Notch4
3.3 Cathepsin S Upregulated TSLP
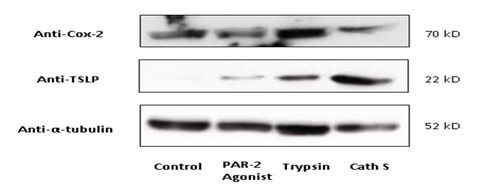
To interrogate whether Cathespin S interacts with protease-activated receptors, HEK293T cells were transfected with pCMV-EGF-PPAR2-FLAG-His plasmids in the presence or absence of Cathepsin S overexpression for a co-immunoprecipitation assay. By pulling down Flag-tagged PAR2 with the anti-FLAG antibody, Cathepsin S was co-precipitated and immunoblotted along with PAR2 (Figure 3). Since Cox2 is a target gene for PAR2- activation [22], we checked if Cathepsin S upregulates Cox2 expression. As shown in Figure 4, trypsin, an agonist for PAR-2 signaling, increased Cox2 expression to a greater extent than that of TSLP expression. In contrast, Cathepsin S and the PAR2 agonist, SLIGRL (a peptide expressed in mice) could not upregulate Cox2 but TSLP, indicating that Cathepsin S-mediated itching may be triggered through the activation of TSLP receptor on peripheral afferent neurons [23].
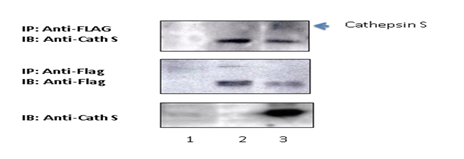
Figure 3:Protease-activated receptor 2 physically interacted with Cathepsin S in HEK293T cells. HEK293T cells
were transfected with pCMV-EGF-PPAR2-FLAG-His plasmids in the presence or absence of Cathepsin S. Cathepsin S was found to co-precipitate with PAR2. Abbreviation? Cath S: Cathepsin S, IP: immunoprecipitation; IB: Immunoblotting; experimental conditions? (1) 293 T control, (2) 293 T + EGF-PPAR2-Flag-His, (3) 293 T + EGF-PPAR2-Flag-His + Cathepsin S treatment.
Figure 4:Cathepsin S upregulated TSLP but not Cox-2. Lysates extracted from primary human keratinocytesthat were treated with PAR-2 agonist (the 2nd lane), trypsin (the 3rd lane) or Cathepsin S (the 4th lane) were blotted for Cox-2 and TSLP protein expression. In comparison with the effect of SLIGRL (a PAR-2 agonist), Cathepsin S markedly triggered TSLP upregulation on normal human epidermal keratinocytes. The effect of Cathepsin S was also differed from that of trypsin by expressing Cox-2 at the basal level. Abbreviation Cath S: Cathepsin S; PAR2: protease-activated receptor 2.
4. Discussion
Radiation-induced cell signaling can be elicited through the activation of protein receptors. Notch signaling, for
example, confers breast cancer resistance upon cell irradiation [22]. Concurrently, radiation stimulates G protein-coupled receptors (GPCRs) and results in inflammation via the activation of transcription factors [23]. In this study, we elucidated a crosstalk mediator of Notch signaling in evoking itch sensation via the stimulation of PAR-2 receptor. As a G protein-coupled receptor, PAR-2 can be cleaved and subsequently activated by proteinases, such as trypsin and Cathepsin S [24,25]. Nevertheless, Cathepsin S cleaves the N-terminus of PAR2, which hence exposes a tethered ligand, KVDGTS, different than that resulting from trypsin cleavage [25]. This may explain the observation that Cathepsin S led to an expression ratio of TSLP to COX2 larger than that caused by the trypsin or the hexapeptide, SLIGRL.
It has been reported TSLP, as a pruritogen and an itch-inducing stimulus, activates dendritic cells to react to infection and tissue stress [26-28]. Radiation-induced expression of TSLP can be resulting from Cathepsin S- or PAR-2- independent pathways, such as JNK, ERK or Mas-related G-protein-coupled receptor signaling [9,29]. Therefore, the findings of this study are limited to the mechanism of TSLP upregulation through Notch-Cathepsin S-PAR2 pathway with regard to a wide panel of radiation-induced itching mediators. As many cytokines and inflammation factors can also mediate radiation-induced itch sensation, the impacts of TSLP upregulation and Notch inhibition on itching using animal models are required to be further evaluated with radiation.
In conclusion, this study demonstrated a new target gene of Notch. Cathepsin S expression was found along with the expression of many other genes, elevated in the Notch4 ICD transgenic mammary tumor. Mechanistically, we identified the role of Notch from the interaction between NICD4 and the CBF binding site of Cathepsin S promoter as well as Cathepsin S-mediated TSLP upregulation in the Notch-Cathepsin S-PAR2-TSLP signaling pathway. This work potentially identified molecular mechanisms by which Notch may induce itching in irradiated skin and may shed a light onto new methods for ameliorating itch sensation.
Acknowledgement
The authors thank Dr. Ethan A Lerner and Dr. Vemuri B Reddy from the Massachusetts General Hospital, Harvard Medical School as well as Dr. William Lowther and Dr. Robert Callahan from the National Cancer Institute of USA for technical support.
Conflict of Interest
The authors declare no conflict of interest.
References
- Salvo N, Barnes E, van Draanen J, et al. Prophylaxis and management of acute radiation-induced skin reactions: a systematic review of the literature. Curr Oncol 17 (2010): 94-112.
- Wang J, Wakeman TP, Latha JD, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells 28 (2010): 17-28.
- Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis 34 (2013): 1420-1430.
- Perry WL, Hustad CM, Swing DA, O’Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet 18 (1998): 143-146.
- Chastagner P, Israël A, Brou C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep 7 (2006): 1147-1153.
- Mazaleyrat SL, Fostier M, Wilkin MB, et al. Down-regulation of notch target gene expression by suppressor of deltex. Dev Biol 255 (2003): 363-372.
- Chen XL, Chinchilla P, Fombonne J, et al. Patched-1 proapoptotic activity is downregulated by modification of k1413 by the e3 ubiquitin-protein ligase itchy homolog. Mol Cell Biol 34 (2014): 3855-3866.
- Seo HR, Bae S, Lee Y-S. Radiation-induced cathepsin S is involved in radioresistance. Int J Cancer 124 (2009): 1794-1801.
- Reddy VB, Sun S, Azimi E, Elmariah SB, Dong X, et al. Redefining the concept of protease-activated receptors: cathepsin S evokes itch via activation of Mrgprs. Nat Commun 6 (2015): 7864.
- Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, et al. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol 174 (2005):1205-1212.
- Roberts R. Lysosomal cysteine proteases: structure, function and inhibition of cathepsins. Drug News Perspect 18 (2005): 605-614.
- Kempkes C, Buddenkotte J, Cevikbas F, Buhl T, Steinhoff M. Role of PAR-2 in Neuroimmune Communication and Itch. In: Carstens E, Akiyama T, eds. Itch: Mechanisms and Treatment. Frontiers in Neuroscience. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. http://www.ncbi.nlm.nih.gov/books/NBK200911/. Accessed December 23, 2015.
- Li JJ. Cancer Stem Cells and Radiotherapy. In: Strauss J, Jr WS, Woloschak GE eds. Breast Cancer Biology for the Radiation Oncologist. Springer; 2015.
- Tripathi R, Rath G, Jawanjal P, et al. Clinical impact of de-regulated Notch-1 and Notch-3 in the development and progression of HPV-associated different histological subtypes of precancerous and cancerous lesions of human uterine cervix. PLoS ONE 9 (2004): e98642.
- Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells 28 (2010): 17-28.
- Sun Y, Klauzinska M, Lake RJ, et al. Trp53 regulates Notch 4 signaling through Mdm2. J Cell Sci 124 (2011): 1067-1076.
- Wang H, Wen S, Bunnett NW, Leduc R, Hollenberg MD, MacNaughton WK. Proteinase-activated receptor-2 induces cyclooxygenase-2 expression through beta-catenin and cyclic AMP-response element-binding protein. J Biol Chem283(2008): 809-815.
- Green D, Dong X. The cell biology of acute itch. J Cell Biol213 (2016): 155-161.
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet 26 (2000): 484-489.
- Sun Y, William Lowther, Katsuaki Kato, et al. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-? signaling. Oncogene 24 (2005): 5365-5374.
- Sun Y, Nonobe E, Kobayashi Y, et al. Characterization and expression of l-amino acid oxidase of mouse milk. J. Biol. Chem 277 (2002): 19080-19086.
- Lagadec C, Vlashi E, Alhiyari Y, Phillips TM, Bochkur Dratver M, Pajonk F. Radiation-induced Notch signaling in breast cancer stem cells. Int J Radiat Oncol Biol Phys 87 (2013): 609-618.
- Sun L, Ye RD. Role of G protein-coupled receptors in inflammation. Acta Pharmacol Sin 33(2012): 342-350.
- Miike S, McWilliam AS, Kita H. Trypsin induces activation and inflammatory mediator release from human eosinophils through protease-activated receptor-2. J Immunol 167 (2001): 6615-6622.
- Elmariah SB, Reddy VB, Lerner EA. Cathepsin s signals via par2 and generates a novel tethered ligand receptor agonist. PLoS ONE 9 (2014): e99702.
- Wilson SR, Thé L, Batia LM, et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell 155 (2013): 285-295.
- Turner MJ, Zhou B. A new itch to scratch for TSLP. Trends in Immunology 35 (2014): 49-50.
- Liu Y-J. TSLP in epithelial cell and dendritic cell cross talk. Adv Immunol 101 (2009): 1-25.
- Jang Y, Jeong SH, Park Y-H, et al. UVB induces HIF-1?-dependent TSLP expression via the JNK and ERK pathways. J Invest Dermatol 133 (2013): 2601-2608.