Gene Expression and Interactome Analysis of Candidate Effectors Associated with Pre- and Post-Haustorial Hemileia vastatrix-Coffee Interaction
Article Information
Isabel Samila Lima Castro1#, Pedro Ricardo Rossi Marques Barreiros1#, Tiago Antônio de Oliveira Mendes2, Juan Carlos Florez1, Edson Mario de Andrade Silva2, Brenda Neves Porto1, Laércio Zambolim1, Eveline Teixeira Caixeta1, 3*
1Instituto de Biotecnologia Aplicada à Agropecuária (BIOAGRO), Universidade Federal de Viçosa, Viçosa, Minas Gerais 36570-900, Brazil
2Departamento de Bioquímica e Biologia Molecular, Universidade Federal de Viçosa, Viçosa, Minas Gerias 36570-900, Brazil
3Brazilian Agricultural Research Corporation (Embrapa), Embrapa Coffee, Brasília 70770-901, Brazil
#ISLC and PRRMB have contributed equally to this work.
*Corresponding author: Eveline Teixeira Caixeta, Institute of Biotechnology Applied to Agriculture (BIOAGRO), Federal University of Viçosa, Viçosa, Minas Gerais 36570900, Brazil.
Received: 24 August 2022; Accepted: 29 August 2022; Published: 13 October 2022
Citation: Isabel Samila Lima Castro, Pedro Ricardo Rossi Marques Barreiros, Tiago Antônio de Oliveira Mendes, Juan Carlos Florez, Edson Mario de Andrade Silva, Brenda Neves Porto, Laércio Zambolim, Eveline Teixeira Caixeta. Gene Expression and Interactome Analysis of Candidate Effectors Associated with Pre- and Post-Haustorial Hemileia Vastatrix-Coffee Interaction Journal of Biotechnology and Biomedicine 5 (2022): 198-213.
View / Download Pdf Share at FacebookAbstract
The present study sought to analyze the putative secreted proteins of Hemileia vastatrix, with potential to function as effector proteins. The H. vastatrix secretome was subjected to functional categorization and the greatest similarities were observed between species of the genus Puccinia sp (398), and Melampsora larici-populina (82). Based on the secretome, 415 Gene Ontology terms were extracted. The putative secretome was also compared to the high-throughput transcriptome of coffee-H. vastatrix interactions. By the transcriptome comparison data and the results of functional annotation and characteristics associated with effector proteins, 15 genes were selected and analyzed using RT-qPCR during compatible and incompatible coffee-H. vastatrix interactions. The expression patterns suggested that the EHv33-18 and EHv33-25 candidate effector may be responsible for faster communication between pathogen and the host during incompatible interaction. Other six candidate are involved in the biotrophic stage of infection, which is characterized by an increase in the expression of effectors, and in enzymes involved in secondary metabolism. Phylogenetic analysis suggest that these eight genes follow evolutionary mechanisms exclusive to the coffee-H. vastatrix interaction, making them important targets in studies aimed at obtaining durable resistance to this disease. Interactomic network made between coffee proteins and H. vastatrix proteins was obtained for the first time and revealed a wide network of interactions between effector EHv33-19 and coffee proteins. The obtained results suggest that there may be communication between the pathogen and the host in the early stage of infection during the urediniospores germination phase. This indicated pre-haustorial resistance complementary to post-haustorial resistance.
Keywords
Plant-pathogen interaction; Secretome; Effector proteins; Gene expression; RT-qPCR; Phylogeny; Interaction network
Plant-pathogen interaction articles Plant-pathogen interaction Research articles Plant-pathogen interaction review articles Plant-pathogen interaction PubMed articles Plant-pathogen interaction PubMed Central articles Plant-pathogen interaction 2023 articles Plant-pathogen interaction 2024 articles Plant-pathogen interaction Scopus articles Plant-pathogen interaction impact factor journals Plant-pathogen interaction Scopus journals Plant-pathogen interaction PubMed journals Plant-pathogen interaction medical journals Plant-pathogen interaction free journals Plant-pathogen interaction best journals Plant-pathogen interaction top journals Plant-pathogen interaction free medical journals Plant-pathogen interaction famous journals Plant-pathogen interaction Google Scholar indexed journals Secretome articles Secretome Research articles Secretome review articles Secretome PubMed articles Secretome PubMed Central articles Secretome 2023 articles Secretome 2024 articles Secretome Scopus articles Secretome impact factor journals Secretome Scopus journals Secretome PubMed journals Secretome medical journals Secretome free journals Secretome best journals Secretome top journals Secretome free medical journals Secretome famous journals Secretome Google Scholar indexed journals Effector proteins articles Effector proteins Research articles Effector proteins review articles Effector proteins PubMed articles Effector proteins PubMed Central articles Effector proteins 2023 articles Effector proteins 2024 articles Effector proteins Scopus articles Effector proteins impact factor journals Effector proteins Scopus journals Effector proteins PubMed journals Effector proteins medical journals Effector proteins free journals Effector proteins best journals Effector proteins top journals Effector proteins free medical journals Effector proteins famous journals Effector proteins Google Scholar indexed journals Gene expression articles Gene expression Research articles Gene expression review articles Gene expression PubMed articles Gene expression PubMed Central articles Gene expression 2023 articles Gene expression 2024 articles Gene expression Scopus articles Gene expression impact factor journals Gene expression Scopus journals Gene expression PubMed journals Gene expression medical journals Gene expression free journals Gene expression best journals Gene expression top journals Gene expression free medical journals Gene expression famous journals Gene expression Google Scholar indexed journals RT-qPCR articles RT-qPCR Research articles RT-qPCR review articles RT-qPCR PubMed articles RT-qPCR PubMed Central articles RT-qPCR 2023 articles RT-qPCR 2024 articles RT-qPCR Scopus articles RT-qPCR impact factor journals RT-qPCR Scopus journals RT-qPCR PubMed journals RT-qPCR medical journals RT-qPCR free journals RT-qPCR best journals RT-qPCR top journals RT-qPCR free medical journals RT-qPCR famous journals RT-qPCR Google Scholar indexed journals Phylogeny articles Phylogeny Research articles Phylogeny review articles Phylogeny PubMed articles Phylogeny PubMed Central articles Phylogeny 2023 articles Phylogeny 2024 articles Phylogeny Scopus articles Phylogeny impact factor journals Phylogeny Scopus journals Phylogeny PubMed journals Phylogeny medical journals Phylogeny free journals Phylogeny best journals Phylogeny top journals Phylogeny free medical journals Phylogeny famous journals Phylogeny Google Scholar indexed journals Interaction network articles Interaction network Research articles Interaction network review articles Interaction network PubMed articles Interaction network PubMed Central articles Interaction network 2023 articles Interaction network 2024 articles Interaction network Scopus articles Interaction network impact factor journals Interaction network Scopus journals Interaction network PubMed journals Interaction network medical journals Interaction network free journals Interaction network best journals Interaction network top journals Interaction network free medical journals Interaction network famous journals Interaction network Google Scholar indexed journals agricultural articles agricultural Research articles agricultural review articles agricultural PubMed articles agricultural PubMed Central articles agricultural 2023 articles agricultural 2024 articles agricultural Scopus articles agricultural impact factor journals agricultural Scopus journals agricultural PubMed journals agricultural medical journals agricultural free journals agricultural best journals agricultural top journals agricultural free medical journals agricultural famous journals agricultural Google Scholar indexed journals pathogen colonization articles pathogen colonization Research articles pathogen colonization review articles pathogen colonization PubMed articles pathogen colonization PubMed Central articles pathogen colonization 2023 articles pathogen colonization 2024 articles pathogen colonization Scopus articles pathogen colonization impact factor journals pathogen colonization Scopus journals pathogen colonization PubMed journals pathogen colonization medical journals pathogen colonization free journals pathogen colonization best journals pathogen colonization top journals pathogen colonization free medical journals pathogen colonization famous journals pathogen colonization Google Scholar indexed journals chromatin articles chromatin Research articles chromatin review articles chromatin PubMed articles chromatin PubMed Central articles chromatin 2023 articles chromatin 2024 articles chromatin Scopus articles chromatin impact factor journals chromatin Scopus journals chromatin PubMed journals chromatin medical journals chromatin free journals chromatin best journals chromatin top journals chromatin free medical journals chromatin famous journals chromatin Google Scholar indexed journals
Article Details
1. Introduction
Coffee (Coffea arabica and C. canephora), an important global agricultural commodity, is mainly affected by leaf rust caused by the fungus Hemileia vastatrix, which belongs to the phylum Basidiomycota, and the order Pucciniales. H. vastatrix is a biotrophic pathogen that infects only coffee plants. The disease causes infected leaves to fall, leading to flower abortion, malformation of grains, and drying of branches, culminating in direct damage to agricultural production [1,2]. Due to recent outbreaks of the disease, which were collectively termed ‘The Big Rust’ by biologist Peter Baker, and involving Latin and Central American countries, H. vastatrix regained its notoriety, becoming a great concern to coffee-producing countries [3-5]. Great losses occurred in Colombia in 2008, leading to a 31% reduction in production. In 2013/2014, the disease caused significant production losses in Central America and Mexico, and in 2014/2015, losses were also reported in Peru and Ecuador [4,5]. The impact caused was similar to that observed in Ceylon in the 19th century, when coffee production was eradicated. In Hawaii, H. vastatrix was reported for the first time in 2020 [6].
Nowadays, H. vastatrix is present in all the coffee growing regions [7]. Despite the efficacy of fungicides, the use of resistant cultivars is the best control method, as it represents an economical, efficient, and sustainable approach. However, a major challenge faced by coffee breeders is the emergence of new races of the pathogen that are capable of overcoming the resistance of otherwise-resistant cultivars. For instance, the resistance of cultivars Oeiras and Icatú Vermelho was overcome approximately 9 and 15 years, respectively, after their release [8]. The main threat to the durability of these cultivars lies in the variability of the pathogen. Over 50 physiological races of H. vastatrix have been identified globally [9]. In Brazil, since 1971, more than 15 of these races have been reported [1,10,11]. This wide genetic variability, which is common to most biotrophic pathogens, has been recognized based on infection patterns observed in a series of 27 coffee clones used to characterize H. vastatrix races [7,12]. This broad variability, continuous emergence of new races, as well as the existence of complex races, demonstrate the high evolutionary potential of H. vastatrix, and represents a great challenge for the development of coffee cultivars with durable resistance [9,10].
Upon interacting with coffee plants, rust-causing fungi secrete a number of effector proteins (also known simply as effectors). Effectors modify the structure and function of host cells, allowing the establishment of pathogen colonization [13]. Some of these effectors, called avirulence proteins (Avr), are recognized by proteins encoded by resistance genes (R genes), triggering defense responses by the plant against the infection [14,15]. Through genetic studies of interactions of the rust (Melampsora lini) and flax [16], Flor (1956) showed that plant-pathogen interactions are determined by genotypes of the host and of the pathogen, in a gene-for-gene interaction. In this way, resistant phenotypes are only observed when the gene of dominant resistance (R) of the host interacts with its respective avirulence gene (Avr) of the pathogen. At least nine dominant genes (SH1 to SH9) that confer resistance to coffee rust have been identified [17,18]. Based on Flor theory, it was thus inferred that there are at least nine Avr genes in H. vastatrix. However, today, it is recognized that there are additional genetic factors which have not yet been identified [2,19,20].
In different pathosystems, mutations in Avr genes allow the pathogen to overcome the resistance conferred by certain genes of the host [21]. Hemileia vastatrix is an obligate haustorium-forming parasite. This structure plays an important role in the biotrophic phase of infection, where it is responsible for absorbing nutrients from the host [22]. Haustoria also induce structural changes in host cells, such as cytoskeletal rearrangement, nucleus migration, and chromatin condensation [23]. These changes are believed to be induced by the action of effectors produced in the haustoria, and which are secreted into the extra-haustorial matrix and translocated into the plant cell. Once within host cells, effectors can alter their metabolism and defense pathways [13]. Plant resistance responses to rusts are usually observed after haustorium formation, indicating that Avr genes are expressed in these structures [24]. Effectors can thus be defined as molecules released by, or associated with, a pathogen, with the ability to modify the physiology of another organism [25]. These molecules have certain common characteristic in that they are small proteins, usually secreted, are rich in cysteine residues, and have specific conserved motifs at the N-terminal region [21]. Another important common characteristic of effectors is that they have a low degree of conservation; that is, it is difficult to determine the functions they serve in the cell, since they have little similarity to other proteins of known function. For this reason, studies aiming to elucidate the mechanisms of action of these molecules are scarce [26].
To improve the knowledge of rust effectors and elucidate pathogen and host interaction, the genome and secreted proteins should be analyzed in detail. Protein-protein interaction networks have, also, emerged as a powerful resource to complement genetic data. The development of protein-protein interaction networks (interactome) help to understand important processes involved in cell function and plant physiology. Since a particular biological function is not commonly attributed to a single molecule but to a functional group of molecules, the interactome analysis can help to understand sets of protein interactions related to a particular function. This strategy also help to elucidate the individual role of proteins in the complex interactions networks, revealing the essentiality of genes and proteins [27].
Interspecific networks protein interaction can be used especially as tools for predicting critical proteins involved in the process of pathogen infection and host resistance, improving plant-pathogen interaction knowledge [28]. Thus, strategies involving biochemistry, genetics, and bioinformatics-often in combination-have been successfully used in the identification of effector protein-coding genes in filamentous fungi [29]. However, few studies have focused on identifying and characterizing effector protein genes in H. vastatrix. The availability of such genes would help to elucidate mutational mechanisms involved in overcoming the coffee resistance, contributing to the development of cultivars with durable resistance. Moreover, knowledge of effector proteins could assist the development of more effective strategies for the control of plant diseases through breeding. In order to obtain a reference genome of H. vastatrix, Porto et al. (2019) sequenced and assembled the genome of the race XXXIII of this species [30]. The obtained genome comprises 576 Mb, with 13,364 predicted genes encoding 13,034 putative proteins. Based on the proteome, the secretome was predicted using a pipeline combining different bioinformatics approaches. Proteins containing signal peptides, and flagged as being related to secretory pathway signal peptides, were pre-selected. Predictions of subcellular localization, and absence of transmembrane domains, were also employed to select the secretome. Next, cysteine residues and characteristic effector motifs, such as [YWF] xC, CxxC and CxxxC [26], were screened for each identified secreted protein. A total of 615 putative secreted proteins was identified. To improve the knowledge of H. vastatrix proteins with potential effector function, in the present work, the fungus genome and putative proteins was analyzed in detail. The focus was the putative secreted proteins of H. vastatrix with the greatest similarities with proteins of other rust fungi (Pucciniales). Approaches of gene expression, phylogeny and construction of protein interaction network (interactome) were used to improve the H. vastatrix-coffee interaction knowledge and to characterize candidate genes able to function as H. vastatrix effector proteins.
2. Materials and Methods
2.1. Functional characterization of H. vastatrix secretome
Functional annotation of the H. vastatrix secretome was carried out using BLAST2GO software [31]. The secretome, which is composed of 615 candidate effector genes, was obtained from the reference genome of H. vastatrix race XXXIII, provided by Porto et al. (2019) [30]. The procedure commenced with a BLASTP search [32] of all 615 contigs of the H. vastatrix secretome against the non-redundant NCBI (National Center for Biotechnology Information) database. Hits with an e-value lower than or equal to 10-10, and with a minimum alignment value [HSP (High Score Pairs) length] of 33 were considered significant. Based on BLASTP results, BLAST2GO extracts terms from Gene Ontology (GO) term enrichments for each contig. The three categories of the electronically designed terms were “Molecular Function”, “Biological Process”, and “Cellular Component”. The last step of BLAST2GO consisted of annotation of Enzyme Codes (EC), and a search for metabolic maps using KEGG (Kyoto Encyclopedia of Genes and Genomes) for the contigs.
2.2. Selection of candidate effector genes for expression analysis
To select candidate genes to be analyzed by RT-qPCR, sequences encoding the 615 secreted proteins [30] were compared with the high-throughput transcriptome of coffee-H. vastatrix interactions [33]. The transcriptome was constructed by inoculation of C. arabica cv. Caturra Vermelho (CIFC 19/1) and Híbrido de Timor (CIFC 832/1) with H. vastatrix race XXXIII, aiming to distinguish compatible and incompatible interactions, respectively. To select candidate genes, we compared the predicted secretome with two transcriptome libraries, which corresponded to 12 hours after infection library (library 12 hai) and 24 hours after infection (library 24 hai), in compatible interactions. We selected: (a) three genes found only in the library 12 hai, and not in the library 24 hai; (b) two genes found only in the library 24 hai; (c) five genes found in both libraries (12 and 24 hai); and (d) five genes with no similarity to any of the transcriptome libraries (not found in the library 12 nether in the library 24). The functional annotation and the characteristics associated with the effector proteins (e.g. small sequence size) of the predicted protein in each library were also considered for candidate gene selection. Thus, based on these criteria, 15 genes were selected. Specific PCR primers for each of the candidate gene sequences were designed (Table 1) using NCBI’s Primer-BLAST software (available at http://www.ncbi. nlm.nih.gov/tools/primer-blast/), with the following settings: amplicon length between 90 and 150 bp, primer size 20 ± 2 bp, annealing temperature (AT) between 55 and 60 ºC, and GC content ± 50%.
2.3. Hemileia vastatrix isolate inoculum
For RT-qPCR, we used the same pathosystem as that employed in previous transcriptome work [33], and also the same fungal isolate of the reference genome [30]. H. vastatrix isolate Hv-02, biologically characterized as race XXXIII [11] was multiplied according to the methodology proposed by Zambolim and Chaves (1974) [34] in seedlings of Caturra coffee (CIFC 19/1).
|
Genes |
NCBI access number(contig) |
Primer sequence (5′→3′) |
Amplicon (pb) |
Efficiency (%) |
|
EHv33-18 |
PHNK01082774 |
F: AGAAATGGCCAAGCCACCTT |
85 |
86.1 |
|
R: TCAGCGATTGAACTACCCCT |
||||
|
EHv33-19 |
PHNK01019218 |
F: GGTGTACTCCCTTTGCACAT |
140 |
83.6 |
|
R: TGGTTGGTTTAGCCCTGTGA |
||||
|
EHv33-20 |
PHNK01004946 |
F: TGGCAAACGGCACTATCACT |
99 |
98.8 |
|
R: TTGCTGTGACCACCCCAAAA |
||||
|
EHv33-21 |
PHNK01019072 |
F: GCAGCGCATTCAAGAACGTA |
114 |
84.7 |
|
R: CAATGGAGCAGCGCAAATCA |
||||
|
EHv33-22 |
PHNK01016829 |
F:TCGGTTGACGATGAACAGGT |
116 |
91.4 |
|
R: TTGCCGCTCTCAATGGTACA |
||||
|
EHv33-23 |
PHNK01023474 |
F: TCTTTCCACAACTTGGCTTGG |
90 |
81.5 |
|
R: ATCAGCATCCACACCCTCATT |
||||
|
EHv33-24 |
PHNK01028242 |
F: TTGGCGAATCAGCTGGGTAA |
145 |
87.6 |
|
R: ATATTTCAGCGCCTGCTGGT |
||||
|
EHv33-25 |
PHNK01059048 |
F: AAGGAGGTTGTGGCCATTTCT |
129 |
90.9 |
|
R: AGACTGCCATTCACTGACCA |
||||
|
EHv33-26 |
PHNK01003126 |
F: TTGCCAACCTTACCGTGACA |
129 |
85.3 |
|
R: ACGTCTGTGCCATTGCTTCA |
||||
|
EHv33-27 |
PHNK01027818 |
F: TCAACTTTGGGGATGCCTCTT |
106 |
96 |
|
R: GGAGTTGTAGTTCAGGATCGGA |
||||
|
EHv33-28 |
PHNK01065933 |
F: AGCGTAATCCTGCCATCCAA |
82 |
92.9 |
|
R: TCCAAACGTCTTACGCAGCA |
||||
|
EHv33-29 |
PHNK01008940 |
F: TGCCATTCAACTTGCTGTTGC |
105 |
107 |
|
R: AATGGTTCCCATCATGAGCAG |
||||
|
EHv33-30 |
PHNK01013702 |
F: TGTCATGACTCTTCCGCCTT |
105 |
97.3 |
|
R: TGCACCACAGAAAGCAGGTA |
||||
|
EHv33-31 |
PHNK01062430 |
F: TGCTGTTGCTCTATGTCGCT |
95 |
98.6 |
|
R: GCAGCCAGGTACCATATCCTAT |
||||
|
EHv33-32 |
PHNK01110291 |
F: TGTCATGACTCTTCCGTCTTCC |
135 |
91.9 |
|
R: TGCACCACAGAAAGCAGGTA |
||||
|
CytIII1 |
F: AGTAGATATGAGTCCCTGACC |
173 |
88.4 |
|
|
R: CACCTTCAGCACTTACATCC |
||||
|
Βtub2 |
F: CTGGTGCCGGAAATAATTGG |
95 |
96.5 |
|
|
R: TCAAGAGAATCACAGCCCTCG |
Table 1: Genes selected for expression analysis. (EHv33) candidate effector genes in Hemileia vastatrix race XXXIII; (F) forward primer sequence; (R) reverse primer sequence. Endogenous genes:1 cytochrome c oxidase subunit III, 2 β-tubulin.
2.4. Plant material and experimentation
Coffee varieties Caturra CIFC 19/01, which is susceptible to the rust, and Híbrido de Timor CIFC 832/1, which is resistant, were used in a trial conducted in a growth chamber with temperature and luminosity set as for obtaining the inoculum. The experiment was set up as a completely randomized design, with three biological replicates for each collection time (hai) using fully expanded young leaves. Collections took place at 12, 24, 48, and 72 hai. The 12 hai sampling was used as a reference for temporal gene expression analyses within each interaction. Inoculation was performed following the methodology mentioned for obtaining inoculum [34]. Samples (leaves) were collected and immediately frozen in liquid N2, then stored in an ultrafreezer at -80ºC until use.
2.5. Hemileia vastatrix RNA extraction and gene expression analysis
The previously frozen leaves were macerated in liquid N2, and approximately 100 mg was used for RNA extraction using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s recommendations. Total RNA was quantified using a Qubit RNA BR assay kit (Life Technologies, Carlsbad, California, EUA) and a NanoDropTM spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, EUA) and RNA integrity was determined by 1.5% agarose gel electrophoresis with ethidium bromide staining. Samples of RNA were stored in an ultrafreezer at -80 ºC until use. cDNA synthesis was carried out with 1 μg of total RNA previously treated with 1 μL DNase for 15 min (50 U/μL, Amplification Grade DNAse I, Invitrogen™, Waltham, Massachusetts, USA). cDNA synthesis was performed using ImProm-II™ Reverse Transcription System RT-PCR kit (Promega, Madison, Wisconsin, EUA), following the manufacturer’s instructions. cDNA was stored at -20 ºC until use. For real-time PCR (RT-qPCR), the SYBR Green I fluorescence detection system (Applied Biosystems, Waltham, Massachusetts, USA) was employed using a 7500 Real Time PCR Systems platform (Applied Biosystems, Waltham, Massachusetts, USA). Each reaction was carried out using 50 ng cDNA, 2 µM of each primer, 5 µL SYBR® Green PCR Master Mix (Applied Biosystems, Waltham, Massachusetts, EUA), and 2 µL sterile water in a final volume of 10 µL. Thermal cycling conditions were 95ºC for 10 min for the initial denaturation, followed by 40 cycles of 95ºC for 15 s and 60ºC for 1 min, ending at 95ºC for 15 s. The melting curve stage was adjusted to standard conditions. Primer efficiency (E) was tested by developing a standard curve of five serial cDNA dilution points (1:5) and calculated based on the slope (a) of the standard curve [E = 10 (-1/a)-1], with Ct values obtained for each dilution. Expression levels of candidate genes were calculated using qBase software [35]. All statistical analyses were undertaken using GraphPad Prism software version 5 (GraphPad Software Inc.; La Jolla, California, USA). To determine significant differences between interactions, unidirectional ANOVA was performed with Dunnett’s test (P < 0.05). Means were compared using Tukey’s test (P < 0.05). Data were normalized using selected constitutive genes of H. vastatrix, CytIII (cytochrome c oxidase subunit III) and βtub (β-tubulin), which had already been validated as normalizers for analysis of H. vastatrix gene expression in planta [36].
2.6. Phylogenetic analysis
Based on gene expression, eight genes considered strong candidate effectors were selected for phylogenetic analysis. The sequences of the selected genes were subjected to the BLASTX algorithm against the H. vastatrix genome, aiming to identify possible copies. Hits with an e-value lower than or equal to 10-10 and coverage greater than 40% were considered significant. Next, the sequences of the candidate effectors were again subjected to BLASTX, employing the same parameters, against all organisms (113) from the FungiDB database [FungiDB: Recursos Genômicos de Fungos e Oomicetos (Genomic Resources of Fungi and Oomycetes)] (Release 38, Jul. 5, 2018). Multi-FASTA files with similar sequences were constructed for each potential effector and used for phylogenetic analyses using MEGA7 software (version 7.0.26) [37]. Sequences were aligned using ClustalW with standard parameters. Phylogenetic trees were obtained using the Maximum Likelihood method, under the following parameters: complete deletion of gaps, Tamura-Nei model [38], with 1000 bootstrap replicates.
2.7. Interactome analysis: Prediction of protein-protein interactions
In order to predict protein-protein interactions between H. vastatrix and coffee, the available genomes of C. canephora [39] and H. vastatrix race XXXIII [30] were submitted to prediction of functional domains with high potential to interact using three independent approaches [40]: PEIMAP, PSIMAP [41] and iPfam database. Initially, sequence alignment methods were used to measure the similarity between the proteins of coffee/pathogen and proteins with mapped domain from the databases. The BLASTP alignment [32] was used for PEIMAP. For PSIMAP, PSI-BLAST alignment was chosen [32] using the database SCOP as subject [42] and Markov's hidden models (HMM) for Pfam [43] implemented in the tool hmmpfam [44]. Alignments with values equal to or greater than 40% identity and 70% coverage for PEIMAP and alignments with e-value less than or equal to 10-4 for PSIMAP and iPfam were considered significant [40]. In predicting protein interactions with PEIMAP, plant and pathogen protein sequences with significant alignment with the database sequences were considered to have the same interaction potential presented by the original database proteins. A reliability value was assigned to each interaction pair based on the experimental method used to predict it [45]. For PSIMAP and iPfam, the significant alignments identified characterized protein domains in plant and pathogen proteins. In iPfam, the interaction reliability value corresponds to the sum of the signature bit scores of the Pfam domains identified in the interaction pair, while in PSIMAP the value corresponds to the inverse of the distance of the structural domains identified in the interaction pair. Once the protein interactions in each bank were predicted, a score of reliability was calculated for each predicted interaction in order to integrate the results of the three methods, considering the independence of each one as previously published [40].
2.8. Hemileia vastatrix membrane protein prediction
In order to obtain a set of interactions with higher potential for occurrence in vivo, a filtering of the total interactions was performed by predicting H. vastatrix secreted or surface proteins. The analysis was performed using four software, TMHMM 2.0 [46], SignalP 4.1 [47], Phobius [46] e WoLF PSORT [48]. TMHMM was used to predict potential transmembrane protein helices using HMM. SinalP was used to predict signal peptide in the amino terminal region of protein sequences by integrating neural networks. Phobius was used to predict both transmembrane proteins and signal peptide in amino acid sequences using HMM. WoLF PSORT was used to predict the potential subcellular localization of proteins, which is based on rules and standards observed by experimental analyzes. The prediction using each software was performed separately, the results were concatenated and the redundancies were eliminated. Thus, the filtered interaction network was obtained.
2.9. Gene ontology analysis and visualization of protein-protein interaction networks
Annotation of coffee protein gene ontology was performed using the AgBase database [49], which assigns biological functions to genes according to ontology. The interactions between H. vastatrix proteins and coffee proteins were visualized using the Cytoscape 3.6.0 software [50]. The ModuLand package was used to identify overlapping protein assemblies (modules) in the interaction network based on the propagation affinity method [51]. The BiNGO package [52] was used to detect enriched functions within ModuLand modules using FDR hypergeometric enrichment analyzes based on gene ontology results.
3. Results
The 615 candidate effector genes predicted in the secretome by Porto et al. (2019) [30] were subjected to functional categorization analysis using the BLAST2GO platform. The greatest similarities were observed between species of the genus Puccinia sp (Puccinia striiformis, Puccinia triticina, Puccinia coronata, and Puccinia sorghi) (398), and Melampsora larici-populina (82). Only 25 sequences exhibited similarity to previously deposited H. vastatrix sequences (Figure 1). In this analysis, 91 sequences did not show similarity (hits) with any sequences deposited in the database.
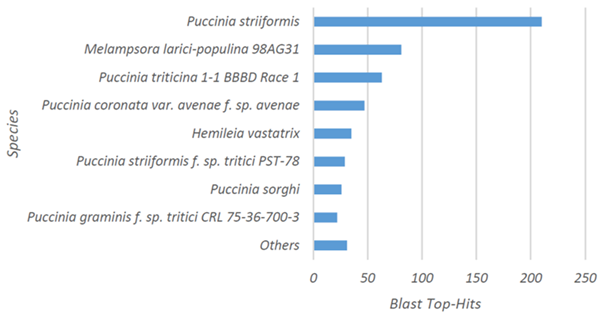
Figure 1: Distribution of species identified at higher frequency in similarity searches using BLASTP for functional categorization and BLAST2GO software. Note the species with highest similarity (Puccinia sp and Melampsora larici-populina) and Hemileia vastatrix.
Based on the secretome, 415 GO terms were extracted and distributed into three categories. For most proteins (496 of 615), it was not possible to obtain GO terms, whereas the remainder (119) presented one or more terms. The most represented category was “Biological Processes”, with 195 terms, followed by “Molecular Function” (118 terms) and “Cellular Component” (102 terms), considering the different hierarchical levels. In the “Biological Processes” category, and considering hierarchical level 3, the most represented terms were “metabolic processes of organic substances”, “primary metabolic processes” and “cellular metabolic processes” (Figure 2). These data agreed with the obtained Enzyme Codes (ECs) (40), in which the most-represented enzyme classes were transferases (20), hydrolases (11) and oxidoreductases (9). For the “Molecular Function” category, the most-frequently represented terms were “transferase activity”, “ion bond” and “hydrolase activity”, considering hierarchical level 3 (Figure 2). In the “Cellular Component” category, the most-represented terms were “intracellular”, followed by “intracellular part” and “membranous organelles” (Figure 2).
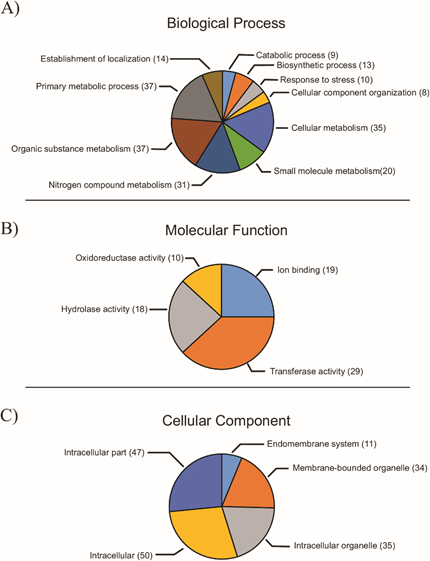
Figure 2: Distribution of GO terms for the a) “Biological Processes”, b) “Molecular Function” and c) “Cellular Component” categories, considering hierarchical level 3.
The 480 proteins with greatest similarities with proteins of other rust fungi (Pucciniales) were analyzed in high-throughput transcriptome of coffee-H. vastatrix interactions. Based on differential expression of the proteins in the transcriptome libraries and functional annotation, 15 genes were selected for temporal expression analysis during H. vastatrix infection (Table 1). A summary of the annotation of the genes selected is in Table 2. Some genes have no GO identity and/or are annotated as hypothetical protein, but they were selected based on the differential expression pattern in the transcriptome data. This strategy was performed because the data showed a low number of potential effectors with GO terms in functional categorization, as well as no similarity with sequences deposited in the database. The small size of the predicted protein was also considered, as it is a characteristic of effectors.
|
Candidate effector |
Description |
GO - ID |
EC |
Library |
Sequence length |
|
EHv33-18 |
Secreted protein |
- |
- |
12/24 hai |
106 |
|
EHv33-19 |
AGC protein kinase |
GO:0016301 |
- |
12 hai |
108 |
|
EHv33-20 |
Hypothetical protein MELLADRAFT_89999 |
- |
- |
- |
343 |
|
EHv33-21 |
ATPase assembly factor ATP10 |
GO:0005739 GO:0007005 GO:0065003 |
- |
- |
182 |
|
EHv33-22 |
Vesicle transporter SEC22 |
GO:0003674 GO:0005783 GO:0005794 GO:0016192 GO:0031410 GO:0032991 GO:0061024 |
- |
12/24 hai |
190 |
|
EHv33-23 |
HECT-domain-containing protein |
GO:0003674 |
- |
24 hai |
257 |
|
EHv33-24 |
Calcium-ion-binding membrane protein |
- |
- |
24 hai |
1006 |
|
EHv33-25 |
- |
- |
- |
- |
140 |
|
EHv33-26 |
Di-copper centre-containing protein |
- |
- |
- |
141 |
|
EHv33-27 |
Hypothetical protein PTTG_29467 |
- |
- |
- |
141 |
|
EHv33-28 |
Hypothetical protein PSHT_06796 |
- |
- |
12/24 hai |
269 |
|
EHv33-29 |
Hypothetical protein PSHT_15882 |
- |
- |
12 hai |
104 |
|
EHv33-30 |
Hypothetical protein PSHT_12651 |
- |
- |
12 hai |
273 |
|
EHv33-31 |
Hypothetical protein PSHT_06796 |
- |
- |
12/24 hai |
148 |
|
EHv33-32 |
Hypothetical protein PSTG_10595 |
- |
- |
12/24 hai |
282 |
Table 2: Summary of the annotation of candidate effector genes selected for gene expression. (GO) Gene Ontology. (EC) Enzyme Code. (Library) Transcriptome library from the compatible H. vastatrix- coffee interaction at 12 and 24 hours after inoculation (hai).
In the Real Time PCR, seven genes (EHv33-18, EHv33-19, EHv33-21, EHv33-27, EHv33-28, EHv33-30 and EHv33-31) showed differential expression between the interactions (incompatible and compatible), and all genes exhibited differences in expression levels over the hai period (Figure 3). The candidate effector genes were collectively grouped (1, 2 and 3) based on similarity in expression patterns (Figure 3), considering infection stage (early or late). The early stage of infection (12 and 24 hai) is considered the pre-haustorial phase, since the initial haustoria are not completely formed, while the late stage of infection (48 and 72 hai) is considered the post-haustorial phase, when such structures are expected to already be formed [33,53,54].
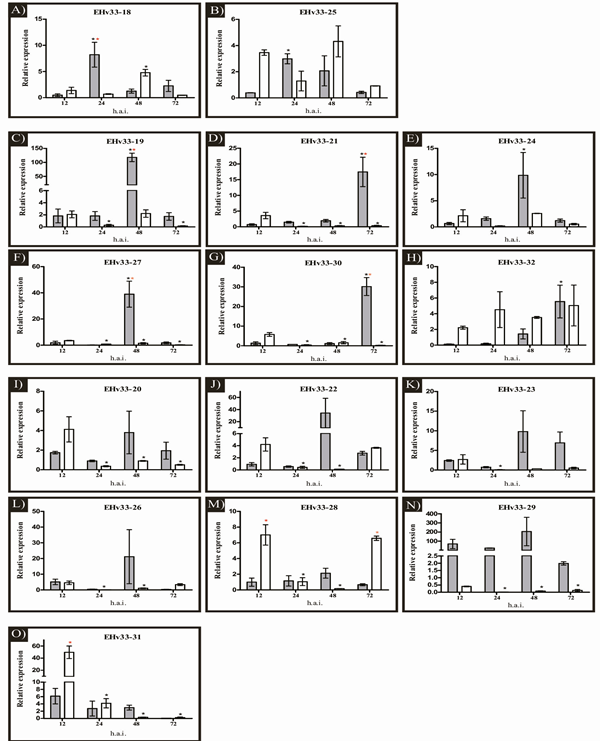
Figure 3: Expression analysis of 15 EHv33 candidate genes encoding secreted proteins in H. vastatrix. Expression patterns were assessed 12, 24, 48 and 72 h after inoculation (hai) of fresh urediniospores into resistant and susceptible plants. The expression levels of target genes were normalized to two endogenous H. vastatrix genes: CytIII and β-tub. (a and b) Genes exhibiting peak expression in the early stage of infection during incompatible interactions; in compatible interactions. Peak expression occurred in the late stage of infection in compatible interactions. (c-h) Genes with peak expression in the late stage of infection during incompatible and compatible interactions. Expression was constant, or showed a peak, in the early stage of infection. (i-o) Genes showing peak expression in the early stage of infection in compatible interactions, and constant expression in incompatible interaction. Error bars = SEM, n = 3 biologically independent replicates. The red asterisk (*) indicates differences in expression levels between the interactions (resistant and susceptible). The black asterisk (*) indicates differences in expression levels within the same interaction, in relation to the 12-hai sample. Dark bars represent the resistant genotype (incompatible interaction). Light bars represent the susceptible genotype (compatible interaction).
Group 1 (Figure 3a-b) consists of two genes (EHv33-18 and EHv33-25), which exhibited peak expression in the early, pre-haustorial, stage of infection in incompatible interactions. In compatible interactions, peak expression was detected in the late, post-haustorial, stage of infection. The EHv33-18 gene demonstrated peak expression at 24 hai in incompatible interactions, while in compatible interactions, it occurred later, at 48 hai. Similarly, for the EHv33-25 gene, peak expression in incompatible interactions also occurred at 24 hai; however, there was no significant difference in expression during infection in compatible interactions. No GO terms were obtained for the genes of this group during functional annotation, which suggested that they are either unknown, or have not been characterized. Although RT-qPCR analysis showed that the EHv33-25 gene was expressed during compatible interactions at 12 and 24 hai, mRNA transcription of this gene was not detected in the transcriptome libraries (Table 2).
Group 2 (Figure 3c-h) comprised EHv33-19, EHv33-21, EHv33-24, EHv33-27, EHv33-30, and EHv33-32 genes. These candidate genes reached peak expression in the late stage (post-haustorial) of infection, during incompatible interactions. For compatible interactions, these genes exhibited constant or decreased expression after 12 hai. For the EHv33-19 and EHv33-27 genes, peak expression occurred at 48 hai in incompatible interactions, while in compatible interactions, expression decreased after 12 hai. Significant differences in expression between the interactions were observed for both genes. For the EHv33-21 and EHv33-30 genes, peak expression was observed at 72 hai in incompatible interactions, and at 12 hai in compatible ones. These two genes also exhibited significant differences between the interactions. For EHv33-24 and EHv33-32, peak expression during incompatible interactions occurred at 48 and 72 hai, respectively. During compatible interactions, there was no significant difference in expression throughout the duration of infection. The EHv33-19, EHv33-30, and EHv33-32 genes showed similarity with sequences from the 12 hai transcriptome library (Table 2), confirming that they were expressed in the early stage of infection in compatible interactions. The EHv33-24 gene showed similarity only with the 24 hai transcriptome library. The EHv33-21 and EHv33-27 genes did not show such similarity with any of the two transcriptome libraries (Table 2).
The EHv33-19 gene was identified with the GO term “kinase activity” in the “Molecular Function” category of GO (GO:0016301). The EHv33-21 gene presented three GO terms: one in the “Cellular Component” category, with the term “mitochondria”, (GO:000573) and two in the “Biological Processes” category, with the terms “mitochondrial organization” (GO:0007005) and “protein-containing complex assembly” (GO:0065003). For the other genes, no GO terms were obtained during functional categorization.
In group 3 (Figure 3i-o) were clustered those candidate effector genes (EHv33-20, EHv33-22, EHv33-23, EHv33-26, EHv33-28, EHv33-29, and EHv33-31) which showed constant expression in incompatible interactions, with peak expression in the early stage (pre-haustorial) of infection, at 12 hai, in compatible interactions. In incompatible interactions, for all genes, an increase in expression was observed at 48 hai, but this peak did not present any statistically significant difference. The EHv33-28 and EHv33-31 genes showed a significant difference between the interactions. Only the candidate effectors EHv33-20 and EHv33-26 did not show similarity with any of the transcriptome libraries (Table 2). In functional annotation, GO terms were obtained for only two candidate effectors in this group. EHv33-22 showed seven GO terms: one in the “Molecular Function” category (GO:0003674), four in the “Cellular Component” category (GO:0005783, GO:0005794, GO:0031410, GO:0032991), and two in the “Biological Processes” category (GO:0016192, GO:0061024). The terms found were “molecular function”, “endoplasmic reticulum”, “Golgi apparatus”, “vesicle-mediated transport”, “cytoplasmic vesicle”, “protein-containing complex”, and “membrane organization”, respectively. The EHv33-23 candidate gene showed one GO term in the “Molecular Function” category (GO:0003674), namely “molecular function”.
Candidate effectors from groups 1 and 2 (EHv33-18, EHv33-19, EHv33-21, EHv33-24, EHv33-25, EHv33-27, EHv33-30, and EHv33-32) were selected for phylogenetic analysis. This approach aimed to determine if these genes produce phylogenetic tree topology profiles similar to that of the Internal Transcribed Spacer (ITS) region (Figure 4a); i.e., if they follow the evolutionary patterns of the analyzed fungal species or exhibit different patterns due to specific selection pressure regarding their functionality. All the eight genes analyzed exhibited tree topology profiles different from that found for the ITS region, located in specific clades of H. vastatrix, and distinct from the other fungi (Figure 4 and ESM 1 and ESM 2).
In order to analyze if these effector candidates have any interaction with coffee proteins, an interactome study was performed by prediction of potential interaction between coffee and H. vastatrix proteins. Considering all coffee proteins, all H. vastatrix proteins and three databases, a protein-protein interaction network with 180,115 predicted interactions was obtained (Table 3 and ESM3). To identify secreted and surface proteins of H. vastatrix genome in the interactome, we used three different software. From the set f 13,034 genes predicted in the H. vastatrix genome, 1,806, 491, 2,386 and 2,203 significant proteins were found by using TMHMM, SignalP, Phobius and WoLF PSORT software, respectively. After concatenating the results, 3,359 H. vastatrix proteins, with no redundancies, were considered secreted or surface proteins. These 3,359 H. vastatrix proteins were used to filter the interactome, resulting in 38,439 interactions between 548 H. vastatrix proteins and 2,382 coffee proteins (ESM4). The filtered interactions network is shown in Figure 5. A total of 16 modules containing proteins with high potential to interact with coffee were obtained, which 15 were functionally signed according to gene ontology analysis. The candidate effectors EHv33-19 was represented in the Filtered Interaction Network, in module 0, showing 333 interactions predicted with coffee proteins. The function associated with module 0 is protein phosphorylation.
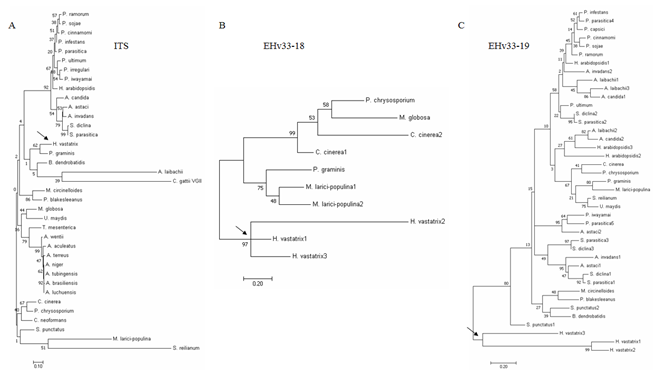
Figure 4: Molecular phylogenetic analysis of the a) ITS and b) of the EHv33-18 and c) EHv33-19 candidate effectors by the Maximum Likelihood method. Bootstrap values are shown next to the branches. Trees are presented on a scale with branch lengths measured according to the number of substitutions per site. Sequence identification codes used to construct the trees, and those of the other effector candidates’ phylogenetic trees, are available in ESM 2 and ESM 1, respectively. Phylogeny was based on alignment of DNA sequences.
|
Method |
Predicted interactions |
H. vastatrix proteins |
Coffee proteins |
|
PEIMAP |
11.812 |
436 |
615 |
|
PSIMAP |
392.145 |
5.522 |
2.693 |
|
iPfam |
248.659 |
2.58 |
8.77 |
|
Total |
180.115 |
2.896 |
3.189 |
Table 3: Protein interaction prediction considering each of the methods used and the total resulting from the concatenation of the data.
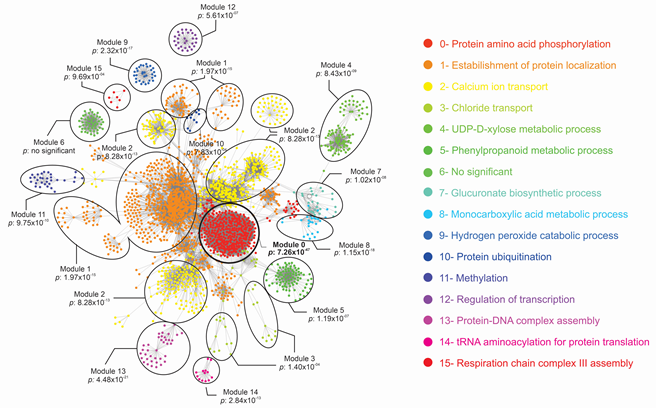
Figure 5: Filtered Protein Interaction Network between H. vastatrix protein and coffee proteins associated with functional interaction modules. Module 0, in which the effector EHv33-19 was represented, is highlighted in bold.
5. Discussion
The similarity found in most candidate effectors of H. vastatrix to Puccinia sp and Melampsora larici-populina proteins can be explained by both rust species having the same biotrophic life styles, belonging to the same phylum (Basidiomycota), and to the same order (Pucciniales). Moreover, Puccinia and Melampsora are model organisms, so they have a large amount of sequence data available in databases. So far, sequences of only 67 H. vastatrix proteins have been deposited in databases, which explains the low similarity observed for proteins of this species. In our work we focused in those protein with similarity to Puccinia and Melampsora. The low number of potential effectors with GO terms (119) extracted by functional categorization, as well as the lack of similarity of some predicted effectors with sequences deposited in the database, indicate that the analyzed genes are neither known, nor characterized. These findings confirm literature reports that describe the low degree of conservation of fungal effector sequences. This low conservation of effectors hinders better understanding of the biological functions, and how such effectors act in the cell during interactions with the host [26]. Furthermore, the lack of similarity may confirm the existence of genes that are specific to the H. vastatrix (race XXXIII)-coffee interaction.
The most represented category, “Biological Processes”, with the main terms “metabolic processes of organic substances”, “primary metabolic process” and “cellular metabolic process” were consistent with the results of metabolic pathway analysis by KEGG, which revealed the presence of coding genes, in particular, of enzymes involved in primary metabolism, and very active metabolic activity. The second most represented category, “Molecular Function” was confirmed by the annotation of ECs, in which the most represented classes were transferases, hydrolases, and oxidoreductases, respectively. The “Cellular Component” category, demonstrates that the activities of effector genes are not restricted to a given cell compartment, but to the entire cell.
The 15 candidate effector genes selected here for gene expression analysis exhibited three different expression patterns. For genes of group 1 (EHv33-18 and EHv33-25), the expression patterns suggest that communication between the pathogen and the host is faster during incompatible interactions. Based on the assumption that the fungus reacts/induces defensive responses in the host, these effectors are possibly involved in fungal attempts to survive the resistance mechanisms of the host plant and/or in the induction of resistance responses. A pre-haustorial resistance response is thus observed, as suggested by Florez et al. (2017) [33] and Castro et al. (2022) [54], unlike post-haustorial resistance, which is usually described for this interaction [55,56]. The EHv33-18 candidate effector may be an important gene for the pathogen during attempts to reestablish infection. The fact that peak expression occurred later, at 48 hai, in compatible interactions, reinforces the idea that pathogen resistance in plants may depend on the rate and extent of synthesis of one or more enzymes induced in the host by the pathogen [57]. Diniz et al. (2012) [57] suggested that the rapid resistance response prevents the formation of haustoria, and this may be the basis for durable resistance. In comparison with the transcriptome data, the EHv33-18 candidate effector showed similarity with reads of the 12 and 24 hai libraries. These libraries belong to the susceptible coffee strains, confirming that this effector is expressed during early stages of compatible interactions. Therefore, these observations reinforce the hypothesis that this effector is important for pathogen infection and is thus expressed in both types of interaction.
For group 2 (EHv33-19, EHv33-21, EHv33-24, EHv33-27, EHv33-30, and EHv33-32) the opposite behavior was observed: candidate effector genes reached peak expression levels in the late stage of infection during incompatible interactions, whereas in compatible interactions, peak expression occurred, in most cases, during the early stage of infection. Guimarães et al. (2015) [58] analyzed the apoplastic fluid of leaves of resistant and susceptible coffee varieties, and also observed that there are two distinct phases of plant defense responses throughout the H. vastatrix infection process: early (24 and 48 hai) and late (72 and 96 hai). These proteins expressed in late stage in resistant genotype may be responsible for activating pathogen genes. At 48 and 72 hai, in susceptible genotypes, all fungal structures were already observed at most infection sites. By contrast, in resistant genotypes, fungal growth stops at different stages, with a higher frequency in the mother-cell stage of the haustorium (MCH), and the formation of fewer haustoria [54,55]. It can thus be inferred that genes from group B are involved in the biotrophic stage of infection, which is characterized by an increase in the expression of effectors, and in enzymes involved in secondary metabolism [59]. These pathogen proteins may act “in combat” against proteins involved in host resistance. The EHv33-27 gene did not present GO terms in functional categorization. This finding suggests that a candidate effector can also be exclusive to the interaction under analysis, which makes it important in studies aimed at understanding the mechanisms of resistance in this pathosystem. The EHv33-19 gene presented the GO term “kinase activity”. Many studies have reported the importance of fungal kinase proteins (KP) during plant infection. The MAPK- (mitogen-activated protein kinase) and the cAMP- (cyclic adenosine monophosphate) dependent protein kinase [protein kinase A (PKA)] are essential for the morphogenesis and virulence of fungi [60-64]. The EHv33-21 gene presented three GO terms: “mitochondria”, “mitochondrial organization” and “protein-containing complex assembly”. The mitochondrial membrane contains important proteins, such as those guiding oxidation reactions in the respiratory chain and ATP synthase. These results are coherent, assuming that the fungus requires more energy in an attempt to overcome host plant resistance.
In Group 3 (EHv33-20, EHv33-22, EHv33-23, EHv33-26, EHv33-28, EHv33-29 and EHv33-31), candidate effector genes reached peak expression during the early stage of infection in compatible interactions. These candidate effectors may be involved in different pathways, which are also important, in addition to being directly related to fungal immunosuppression mechanisms. Most genes in this group showed a decline in expression in the hours subsequent to infection; however, for the EHv33-31 gene, this decrease was more evident, which indicates its importance in the first hours post-infection. This effector may be crucial during the stage of penetration by the fungus into the host, as it is related to differentiation of the fungal structure. Because this expression pattern was only observed in compatible interactions, with constant expression in incompatible interactions, it is also reasonable to infer that the fungus reduces the expression of those genes throughout the infection in an attempt to conserve energy, since the resistance mechanisms of the host had already been overcome.
The genes from groups 1 and 2 were considered strong candidate effectors and were thus selected for phylogenetic analysis. For all these genes, tree topologies were different from the fungal ITS marker tree, suggesting specific evolution. Specific evolution (unlike the rest of the genome) may be related to the selection pressure to which these genes are subjected to due to the high specificity of such interactions. Most candidate effector genes showed more than one genomic copy. This observation is common in species-specific interactions, in which a gene copy usually maintains the basic function of the progenitor gene. In this way, other copies are available to evolve [65]. Thus, our results suggest that such genes are under selective pressure, resulting in H. vastatrix genes being clustered in a single clade, unlike as expected, based on the evolutionary model of the ITS marker. Thus, these results confirm that genes in groups A and B are strong candidate effectors of H. vastatrix.
The interactome between coffee and H. vastatrix proteins revealed a complex network involving 38,439 predicted interactions. To obtain this data, only surface and secreted proteins of the fungus were considered. Since the process of infection occurs with the development of fungal structures and protein secretion in plant tissue, surface proteins and those secreted by the fungus are more exposed to molecular interaction. Among the 15 functional interaction modules obtained, there are important pathways linked to plant defense against pathogen attacks, such as protein phosphorylation and ubiquitination processes, Ca2 + transport, phenylpropanoids and hydrogen peroxide metabolic pathway. The pathways associated with modules 0, 2, 5, 9 and 10 have already been related to plant responses to pathogen attack in previous studies. For example, protein phosphorylation and ubiquitination are post-translational modifications involved in pathogen recognition pathways, signaling and regulation of plant defense responses [66]. The candidate effector EHv33-19 was identified in module 0 of the interactome, suggesting its function during pathogen infection. A high number of interactions with coffee proteins were observed, which may also suggest a critical role during plant defense response. It has been shown that the protein-protein interactions play essential roles for many physiological, pathological and developmental processes in essentially all organisms [67].
5. Conclusion
Interaction network, phylogeny study and gene expression analysis suggest that the genes EHv33-18, EHv33-19, EHv33-21, EHv33-24, EHv33-25, EHv33-27, EHv33-30, and EHv33-32 are strong candidate effectors that may play essential functions during H. vastatrix-coffee interactions, influencing plant resistance mechanisms. The results showed that there may be communication between the pathogen and the host in the early stage of infection during the urediniospores germination phase. This suggests pre-haustorial resistance complementary to post-haustorial resistance, as described previously for the same interactions. The construction and evaluation of the Protein Interaction Network between H. vastatrix and coffee provided, for the first time, a set of interactions with great potential to participate in the plant resistance process. The generated data may shed light on molecular mechanisms of pathogen recognition and inhibition by the plant, as well as having the potential to indicate bioprospective molecules for coffee rust control. Functional biological studies are warranted in order to determine the true function of these effectors during host-pathogen interactions. The present results contribute to better understanding of the molecular mechanisms involved in overcoming resistance by new fungal races, underpinning the development of cultivars with durable resistance. The information obtained in the current study is unprecedented and represents a major step to overcome this challenge.
Acknowledgments
This work was financially supported by the Brazilian Consortium for the Research and Development of Coffee (CBP&D/Café); the Research Foundation of Minas Gerais State (FAPEMIG); the National Research Council (CNPq); the National Institute of Science and Technology (INCT Café) and the Coordination of Improvement of Higher Education Personnel (CAPES).
References
- Zambolim L. Current status and management of coffee leaf rust in Brazil. Tropical Plant Pathology 41 (2016): 1-8.
- Zambolim L, Caixeta ET. An overview of physiological specialization of coffee leaf rust-new designation of pathotypes. International Journal of Current Research 13 (2021): 15564-15575.
- Baker P. The ‘Big Rust’: an update on the coffee leaf rust situation. Coffee Cocoa International 40 (2014): 37-39.
- Avelino J, Cristancho M, Georgiou S, et al. The coffee rust crises in Colombia and Central America (2008-2013): impacts, plausible causes and proposed solutions. Food Security 7 (2015): 303-321.
- McCook S, Vandermeer J. The Big Rust and the Red Queen: Long-term perspectives on coffee rust research. Phytopathology 105 (2015): 1164-1173.
- Keith LM, Sugiyama LS, Brill E, et al. First report of coffee leaf rust caused by Hemileia vastatrix on Coffee (Coffea arabica) in Hawaii. Plant Disease 34 (2022): 2-4.
- Silva MDC, Guerra-Guimarães L, Diniz I, et al. An overview of the mechanisms involved in Coffee-Hemileia vastatrix interactions: Plant and pathogen perspectives. Agronomy 12 (2022): 326.
- Capucho AS, Caixeta ET, Zambolim EM, et al. Herança da resistência do Híbrido de Timor UFV 443-03 à ferrugem-do-cafeeiro. Inheritance of coffee leaf rust resistance in Timor Hybrid UFV 443-03. Pesquisa agropecuária brasileira, Brasília 44 (2009): 276-282.
- Várzea VMP, Marques D. Durable Resistance to Coffee Leaf Rust. In: Zambolim L, Zambolim EM, Várzea VMP, editors. Durable Resistance to Coffee Leaf Rust. UFV. Viçosa (2005): 53-74.
- Cabral PGC, Zambolim EM, Zambolim L, et al. Identification of a new race of Hemileia vastatrix in Brazil. Australasian Plant Disease Notes 4 (2009): 129-130.
- Capucho AS, Zambolim EM, Freitas RL, et al. Identification of race XXXIII of Hemileia vastatrix on Coffea arabica Catimor derivatives in Brazil. Australasian Plant Disease Notes 7 (2012): 189-191.
- Talhinhas P, Batista D, Diniz I, et al. The coffee leaf rust pathogen Hemileia vastatrix: one and a half centuries around the tropics. Molecular Plant Pathology 18 (2017): 1039-1051.
- Zhang S, Li C, Si J, et al. Action mechanisms of effectors in plant-pathogen interaction. International Journal of Molecular Sciences 23 (2022): 6758.
- Jones JDG, Dangl JL. The plant immune system. Nature 444 (2006): 323-329.
- Lang J, Colcombet J. Sustained Incompatibility between MAPK signaling and pathogen effectors. International Journal of Molecular Sciences 21 (2020): 1-26.
- Flor HH. The complementary genic systems in flax and flax rust. Advances in Genetics 8 (1956): 29-54.
- Rodrigues CJ, Bettencourt AJ, Rijo L. Races of the pathogen and resistance to coffee rust. Annual Reviews of Phytopathology 13 (1975): 49-70.
- Bettencourt A, Rodrigues Jr C. Principles and practice of coffee breeding for resistance to rust and other diseases. In: CLARKE RJ, MACRAE R, editors. Coffee Agronomy. London and New York: Elsevier Applied Science Publishers LTD (1988): 199-234.
- Barka GD, Caixeta ET, de Almeida RF, et al. Differential expression of molecular rust resistance components have distinctive profiles in Coffea arabica - Hemileia vastatrix interactions. European Journal of Plant Pathology 149 (2017): 543-561.
- Silva RA da. Caracterização de raças fisiológicas e análise de proteínas candidatas a efetoras em população de Hemileia vastatrix no Brasil. Universidade Federal de Viçosa (2017).
- Stergiopoulos I, de Wit PJGM. Fungal Effector Proteins. Annual Reviews Phytopathology 47 (2009): 233-263.
- Fei W, Liu Y. Biotrophic Fungal Pathogens: a Critical Overview. Applied Biochemistry and Biotechnology (2022).
- Mendgen K, Hahn M. Plant infection and the establishment of fungal biotrophy. Trends in Plant Science 7 (2002): 352-356.
- Dodds PN, Rafiqi M, Gan PHP, et al. Effectors of biotrophic fungi and oomycetes: pathogenicity factors and triggers of host resistance. New Phytologist 183 (2009): 993-1000.
- Dalio RJD. Efetores nas interações planta-patógenos. Revisão Anual de Patologia de Plantas. Passo Fundo 22 (2014): 25-68.
- Duplessis S, Cuomo CA, Lin Y-C, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proceedings of the National Academy of Sciences of the USA 108 (2011): 9166-9171.
- Adadi R, Volkmer B, Milo R, et al. Prediction of Microbial Growth Rate versus Biomass Yield by a Metabolic Network with Kinetic Parameters. PLoS Computational Biology 8 (2012): 1002575.
- Javed Iqbal M, Majeed M, Humayun M, et al. Proteomic profiling and the predicted interactome of host proteins in compatible and incompatible interactions between soybean and Fusarium virguliforme. Applied Biochemistry and Biotechnology 180 (2010): 1657-1674.
- Ellis JG, Rafiqi M, Gan P, et al. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr Current Opinion in Plant Biology 12 (2009): 399-405.
- Porto BN, Caixeta ET, Mathioni SM, et al. Genome sequencing and transcript analysis of Hemileia vastatrix reveal expression dynamics of candidate effectors dependent on host compatibility. PLoS One 14 (2019): e0215598.
- Conesa A, Götz S, García-Gómez JM, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(2005): 3674-3676.
- Altschul SF, Gish W, Pennsylvania T, et al. Basic Local Alignment Search Tool 2 Department of Computer Science. Journal of Molecular Biology 215 (1990): 403-410.
- Florez JC, Mofatto LS, do Livramento Freitas-Lopes R, et al. High throughput transcriptome analysis of coffee reveals pre haustorial resistance in response to Hemileia vastatrix infection. Plant Molecular Biology 95 (2017): 607-623.
- Zambolim L, Chaves GM. Efeito de baixas temperaturas e do binomio temperatura-umidade relativa sobre a viabilidade dos uredosporos de Hemileia vastatrix Berk. et Br. e Uromyces phaseoli typica Arth. Experientiae 17 (1974): 151-184.
- Hellemans J, Mortier G, De Paepe A, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 8 (2007): R19.
- Vieira A, Talhinhas P, Loureiro A, et al. Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biology 115 (2011): 891-901.
- Kumar S, Stecher G, Tamura K, et al. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33 (2016): 1870-1874.
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10 (1993): 512-526.
- Denoeud F, Carretero-Paulet L, Dereeper A, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345 (2014): 1181-1184.
- Flórez AF, Park D, Bhak J, et al. Protein network prediction and topological analysis in Leishmania major as a tool for drug target selection. BMC Bioinformatics 11 (2010): 484.
- Park D, Lee S, Bolser D, et al. Comparative interactomics analysis of protein family interaction networks using PSIMAP (protein structural interactome map). Bioinformatics 21 (2005): 305-333.
- Conte L Lo, Ailey B, Hubbard TJP, et al. SCOP: a structural classification of proteins database. Nucleic Acids Research 28 (2000): 257-259.
- Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research 44 (2016): D279-D285.
- Choo KH, Tong JC, Zhang L. Recent applications of Hidden Markov Models in computational biology. Genomics Proteomics Bioinformatics 2 (2004): 84-96.
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research 39 (2011): 561-568.
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings of the International Conference of Intelligent Systems for Molecular Biology 6 (1998): 175-182.
- Petersen TN, Brunak S, von Heijne G, et al. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8 (2011): 785-786.
- Horton P, Park K-J, Obayashi T, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35 (2007): W585-587.
- McCarthy FM, Wang N, Magee GB, et al. AgBase: a functional genomics resource for agriculture. BMC Genomics 7 (2006): 229.
- Saito R, Smoot ME, Ono K, et al. A travel guide to Cytoscape plugins. Nature Methods 9 (2012): 1069-1076.
- Kovács IA, Palotai R, Szalay MS, et al. Community landscapes: an integrative approach to determine overlapping network module hierarchy, identify key nodes and predict network dynamics. PLoS ONE 5 (2010):e12528.
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 21 (2005): 3448-3449.
- Silva M do C, Várzea V, Guerra-Guimarães L, et al. Coffee resistance to the main diseases: leaf rust and coffee berry disease. Brazilian Journal of Plant Physiology 18 (2006): 119-147.
- Castro ISL, Freitas-Lopes RDL, Ferreira S de S, et al. Transcriptome analysis uncovers the gene expression profile of Hemileia vastatrix (race XXXIII) during the interactions with resistant and susceptible coffee. Agronomy 12 (2022): 444.
- Silva MC, Nicole M, Guerra-Guimarães L, et al. Hypersensitive cell death and post-haustorial defence responses arrest the orange rust (Hemileia vastatrix) growth in resistant coffee leaves. Physiological and Molecular Plant Pathology 60 (2002): 169-183.
- Ramiro DA, Escoute J, Petitot AS, et al. Biphasic haustorial differentiation of coffee rust (Hemileia vastatrix race II) associated with defence responses in resistant and susceptible coffee cultivars. Plant Pathology 58 (2009): 944-955.
- Diniz I, Talhinhas P, Azinheira HG, et al. Cellular and molecular analyses of coffee resistance to Hemileia vastatrix and nonhost resistance to Uromyces vignae in the resistance-donor genotype HDT832/2. European Journal Plant Pathology 133 (2012): 141-157.
- Guerra-Guimarães L, Tenente R, Pinheiro C, et al. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Frontiers in Plant Science 6 (2015): 1-16.
- O’connell RJ, Thon MR, Hacquard S, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics 44 (2012): 1060-1065.
- Banuett F, Herskowitz I. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes & Development 8 (1994): 1367-1378.
- Gold S, Duncan G, Barrett K, et al. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes & Development 8 (1994): 2805-2816.
- Gold SE, Brogdon SM, Mayorga ME, et al. The Ustiago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell 9 (1997): 1585-1594.
- Mitchell’ TK, Dean’ RA. The camp-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. The Plant Cell 7 (1995): 1869-1878.
- Xu J-R, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation, and pathogemc growth in the rice blast fungus Magnaporthe grisea. Genes & Development 10 (1996): 2696-2706.
- Hughes AL. Gene duplication and the origin of novel proteins. Proceedings of the National Academy of Sciences of the USA 102 (2005): 8791-8792.
- Withers J, Dong X. Post-translational regulation of plant immunity. Current Opinion in Plant Biology 38 (2017): 124-132.
- von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417 (2002): 399-403.
