Bioinformatics Analysis Identifies NDRG1 Gene Variants that may be Clinically Relevant
Article Information
Borré, GB1. Pimenta, AT1. Chmieleski, GS1. Moyses, GR1. Sousa, SCB1. Rabi, LT1. Peres, KC1 . Teixeira, ES1. Bufalo, NE1,2,3. Ward, LS1*
1Laboratory of Cancer Molecular Genetics, School of Medical Sciences, University of Campinas (UNICAMP), Campinas/SP, Brazil
2Department of Medicine, São Leopoldo Mandic and Research Center, Campinas/SP, Brazil
3Department of Medicine, Max Planck University Center, Campinas/SP, Brazil
*Corresponding Author: Laura Sterian Ward, Laboratory of Cancer Molecular Genetics, School of Medical Sciences, PO Box 6111, Campinas, SP-Brazil
Received: 28 November 2022; Accepted: 5 December 2022; Published: 14 December 2022
Citation:
Borré GB, Pimenta AT, Chmieleski GS, Moyses GR, Sousa SCB, Rabi LT, Peres KC, Teixeira ES, Bufalo NE, Ward LS. Bioinformatics Analysis Identifies NDRG1 Gene Variants that may be Clinically Relevant. Journal of Bioinformatics and Systems Biology 5 (2022): 158-162.
View / Download Pdf Share at FacebookAbstract
Background: The search of single nucleotide variants that might have the capacity to alter genetic information and influence in regular cellular pathways, enhancing expansion, mitosis and evasion capacity to neoplasm cells, is central in understanding the molecular nature of distinct cellular growth abnormalities and is critical because it might expose new possibilities for therapeutic targets. The expression of NDRG1 protein, encoded by NDRG1 gene, has already been correlated with tumor progression and evasion, but information on different types of neoplasm is still contentious.
Objective: To explore probable correlations of susceptibleness, progression and clinical characteristics between NDRG1 gene polymorphisms (SNPs) and patients that developed thyroid tumors.
Methods: SNPs were obtained from the NCBI dbSNP. The encoded protein primary sequences were got from the UniProt database. We employed the three FASTA primary sequences to analyze the amino acid changes. The bioinformatics tools used were: PredictSNP1.0 (which encompasses: PANTHER, SNAP, PolyPhen-1, PhD-SNP, nsSNPAnalyze, SIFT, PredictSNP, PolyPhen-2, MAPP,); I-Mutant2.0; MUpro; PROVEAN; Haploview and SNPs3D).
Results: The NCB database reports 319 missense SNPs in the NDRG1 gene. The SIFT tool predicted that 51 nsSNPs of 109 (which means 46.78%) were deleterious; the SNAP tool predicted nearly 30%; PolyPhen-2, 53 (48.62%); 52 (47.70%) derived from PhD-SNP; PolyPhen-1 indicated 38 nsSNPs (approximately 35%); and MAPP showed 47 (which is 43%). Finally, the PredictSNP toll contemplated 13 (approximately 12%) nsSNPs deleterious by all integrated tools, including rs201348291 and rs15132213, whose scores were the most significant, thus indicating a higher possibility that these SNPs are correlated and infl
Keywords
NDRG1, Thyroid, Bioinformatics, In silico analysis.
NDRG1 articles; Thyroid articles; Bioinformatics articles; In silico analysis articles
NDRG1 articles NDRG1 Research articles NDRG1 review articles NDRG1 PubMed articles NDRG1 PubMed Central articles NDRG1 2023 articles NDRG1 2024 articles NDRG1 Scopus articles NDRG1 impact factor journals NDRG1 Scopus journals NDRG1 PubMed journals NDRG1 medical journals NDRG1 free journals NDRG1 best journals NDRG1 top journals NDRG1 free medical journals NDRG1 famous journals NDRG1 Google Scholar indexed journals Thyroid articles Thyroid Research articles Thyroid review articles Thyroid PubMed articles Thyroid PubMed Central articles Thyroid 2023 articles Thyroid 2024 articles Thyroid Scopus articles Thyroid impact factor journals Thyroid Scopus journals Thyroid PubMed journals Thyroid medical journals Thyroid free journals Thyroid best journals Thyroid top journals Thyroid free medical journals Thyroid famous journals Thyroid Google Scholar indexed journals Bioinformatics articles Bioinformatics Research articles Bioinformatics review articles Bioinformatics PubMed articles Bioinformatics PubMed Central articles Bioinformatics 2023 articles Bioinformatics 2024 articles Bioinformatics Scopus articles Bioinformatics impact factor journals Bioinformatics Scopus journals Bioinformatics PubMed journals Bioinformatics medical journals Bioinformatics free journals Bioinformatics best journals Bioinformatics top journals Bioinformatics free medical journals Bioinformatics famous journals Bioinformatics Google Scholar indexed journals In silico analysis articles In silico analysis Research articles In silico analysis review articles In silico analysis PubMed articles In silico analysis PubMed Central articles In silico analysis 2023 articles In silico analysis 2024 articles In silico analysis Scopus articles In silico analysis impact factor journals In silico analysis Scopus journals In silico analysis PubMed journals In silico analysis medical journals In silico analysis free journals In silico analysis best journals In silico analysis top journals In silico analysis free medical journals In silico analysis famous journals In silico analysis Google Scholar indexed journals DNA articles DNA Research articles DNA review articles DNA PubMed articles DNA PubMed Central articles DNA 2023 articles DNA 2024 articles DNA Scopus articles DNA impact factor journals DNA Scopus journals DNA PubMed journals DNA medical journals DNA free journals DNA best journals DNA top journals DNA free medical journals DNA famous journals DNA Google Scholar indexed journals Nonsynonymous SNPs articles Nonsynonymous SNPs Research articles Nonsynonymous SNPs review articles Nonsynonymous SNPs PubMed articles Nonsynonymous SNPs PubMed Central articles Nonsynonymous SNPs 2023 articles Nonsynonymous SNPs 2024 articles Nonsynonymous SNPs Scopus articles Nonsynonymous SNPs impact factor journals Nonsynonymous SNPs Scopus journals Nonsynonymous SNPs PubMed journals Nonsynonymous SNPs medical journals Nonsynonymous SNPs free journals Nonsynonymous SNPs best journals Nonsynonymous SNPs top journals Nonsynonymous SNPs free medical journals Nonsynonymous SNPs famous journals Nonsynonymous SNPs Google Scholar indexed journals gene articles gene Research articles gene review articles gene PubMed articles gene PubMed Central articles gene 2023 articles gene 2024 articles gene Scopus articles gene impact factor journals gene Scopus journals gene PubMed journals gene medical journals gene free journals gene best journals gene top journals gene free medical journals gene famous journals gene Google Scholar indexed journals EMT articles EMT Research articles EMT review articles EMT PubMed articles EMT PubMed Central articles EMT 2023 articles EMT 2024 articles EMT Scopus articles EMT impact factor journals EMT Scopus journals EMT PubMed journals EMT medical journals EMT free journals EMT best journals EMT top journals EMT free medical journals EMT famous journals EMT Google Scholar indexed journals hepatocarcinoma articles hepatocarcinoma Research articles hepatocarcinoma review articles hepatocarcinoma PubMed articles hepatocarcinoma PubMed Central articles hepatocarcinoma 2023 articles hepatocarcinoma 2024 articles hepatocarcinoma Scopus articles hepatocarcinoma impact factor journals hepatocarcinoma Scopus journals hepatocarcinoma PubMed journals hepatocarcinoma medical journals hepatocarcinoma free journals hepatocarcinoma best journals hepatocarcinoma top journals hepatocarcinoma free medical journals hepatocarcinoma famous journals hepatocarcinoma Google Scholar indexed journals
Article Details
1. Introduction
Background:
The majority of the modifications in human genetic material are called Single Nucleotide Polymorphisms (SNPs) and occur at a single nucleotide base in DNA (1). These variations can be responsible for mutations and modify proteins regarding its function, structure and stability, therefore resulting in different disorders. SNPs can be useful in clinical practice as markers of diseases. They can help identify whether someone has a higher risk of developing a determined disease, also predicting its possible course, the patient's response to medications, and therefore his prognosis. SNPs can help identify the individual response to certain agents such as environmental toxins, in addition to drugs, individualizing a therapeutic response profile and assisting in patient management (3). Nonsynonymous SNPs - nsSNPs or missense variations might alter the translation and transcription process and modify the amino acid residues sequence (4) . Missense variations can destabilize and modify protein solubility and therefore cause malfunction and gene deregulation (4).
The NDRG1 protein is encoded by the NDRG1 gene, which responds to cellular stress signals (6, 7). NDRG1 participates in important cell signaling pathways and is associated with suppression of cell growth and differentiation (8,10,13). However, its role in tumor development and evolution is still controversial. It might have an anti-metastatic effect in some tumors, but, on the contrary, it has been correlated with more undifferentiated tumor, less benign prognosis and higher potential to more aggressiveness and metastasis (10, 11). In colorectal, breast and prostate neoplasms, the expression of NDRG1 presents with lower incidence of metastasis and progression (8,10). In contrast, it correlates with more aggressive prognosis in hepatocarcinoma (12,13). NDRG1 protein is likely to be involved in tumor development and evasion mechanisms through MMP-9 and MMP-2 whereas its effect on the metastatic potential appears to be associated with EMT and E-cadherin activities and also with aberrant methylation (8).
There is evidence of higher expression of the protein in thyroid carcinomas, primary and metastatic, in comparison to normal and benign thyroid affections in studies considering immunohistochemical (10). In patients with thyroid carcinoma its expression was correlated with an higher-risk AMES category and also more advanced TNM stages, suggesting it has a role in thyroid tumor progression (10). Furthermore, p.G136D is a novel carcinoma somatic alteration of NDRG1 that was found in a patient with PTC (33). An exome sequencing study proposed that variants of NDRG1 might have an important role in papillary thyroid cancer (PTC) development and can be possibly used as markers of higher aggressiveness and as prognostic indicators (31). Nonetheless, this gene's role in thyroid pathology is still very little understood.
This study aimed to investigate nsSNPs of the NDRG1 gene that could produce deleterious changes in the protein and could be associated with the carcinogenic process of the thyroid.
2. Materials and Methods
2.1 Data Selection:
he SNPs for this study were selected from the NCBI database (https://www.ncbi.nlm.nih.gov/snp/?term=NDRG1). All three found FASTA sequences, three primary sequences of the proteins encoded by the gene, were contemplated for further analysis, which were taken from the UniProt database (https://www.uniprot.org/uniprot/Q92597) under the code Q92597.
2.2 Data analysis:
The previously described bioinformatics instruments were used to decode and express the effects that the NDRG1 protein suffered from the DNA changes. These effects were analyzed through the following tools:
2.3 Deleterious nsSNPs prediction:
PredictSNP1.0 (http://loschmidt.chemi.muni.cz/predictsnp1/) (15) was used to predict the impact on protein function and structure. This tool allows access to nine top performing prediction tools: PolyPhen-2, SIFT, PhD-SNP, MAPP, PolyPhen-1, PredictSNP and SNAP. PolyPhen-1 (Polymorphism Phenotyping) evaluates the possibility of impact of amino acid substitutions and PolyPhen-2 (Polymorphism Phenotyping v2) estimates the potential effect of an amino acid substitution on the structure and function of a human protein. SIFT (Sorting Intolerant from Tolerant) analyzes if an amino acid substitution modifies protein function (16). SNAP (Screening for Unacceptable Polymorphisms) is a neural network-based method used to predict functional effects of nsSNPs using information from in silico-derived proteins (19) MAPP (Multivariate Analysis of Protein Polymorphism) investigates the physicochemical variation present in each column of a protein sequence alignment and predicts the impact of amino acid substitutions on protein function (17). PhD-SNP (Predictor of Human Deleterious Single Nucleotide Polymorphisms) classifies nsSNPs that change the human DNA causing disease (18). PredictSNP1.0 shows the confidence scores generated by each tool and a consensus prediction as percentages (15).
2.4 Prediction of linkage disequilibrium:
Pairs of altered alleles contained in genetic information can be more likely to be biologically inherited than other alleles from other pairs of SNPs. This is linkage equilibrium. The Haploview software informs the linkage disequilibrium prediction of the human genome (20).
2.5 Protein-protein interaction analysis:
STRING (https://string-db.org/cgi/about) predicts and informs protein-protein interactions. These interactions can be physical or functional.
2.6 Probability analysis of the impact of nsSNPs on the function of the studied protein:
The SNPs3D tool (http://www.SNPs3D.org) classifies and relates which genes can be considered to be involved in specific diseases. The tool also informs whether the nsSNPs modify protein function. The information derives from the analysis of the protein structure and stability and also of sequence conservation.
3. Results
3.1 SNP dataset:
Data on the NDRG1 gene SNPs (NCBI Gene ID: 10397) were retrieved during September 2021 from the NCBI database (https://www.ncbi.nlm.nih.gov/snp/?term=ndrg1), from where 16,383 SNPs were obtained. The 319 that were missense were further evaluated.
3.2 Prediction of deleterious nsSNPs:
147 out of the 319 missense polymorphisms obtained from dbSNP showed amino acid changes and some even had more than one change, so the three FASTA primary sequences were used to analyze these amino acid variations. They correspond to the three protein isoforms, are encoded by the gene in question and were taken from the UniProt database under the code Q92597.
SIFT showed that 51 out of 109 (46.78%) SNPs were deleterious; SNAP indicated 32 (29.35%) deleterious nsSNPs; PolyPhen-2 indicated 53 (48.62%) nsSNPs; PhD-SNP indicated 52 (47.70%) nsSNPs; PolyPhen-1 identified 38 (34.86%) nsSNPs; and MAPP indicated 47 (43.11%) deleterious nsSNPs. The PredictSNP tool analysis considered 13 (11.92%) nsSNPs deleterious by all the integrated tools, as shown in table 1.
Four SNPs (rs2272646, rs3779941, rs3088599 and rs2977497) from intronic regions found in literature were also analyzed, but were found to be mostly neutral.
Extensive literature search was performed for each of the deleterious nsSNPs selected by these tools, but we were unable to find any description or mention of any of them connected to thyroid nodules or thyroid tumors. Unfortunately, all these SNPs had very minor allele frequency (MAF < 0.01).
Table 1: NDRG1 nsSNPs considered deleterious by all the integrated tools
|
nsSNPS |
AA Change |
FASTA |
PredictSNP |
SIFT |
PhD-SNP |
PolyPhen-1 |
PolyPhen-2 |
MAPP |
SNAP |
|
rs142426003 |
Y62C |
1 |
D |
D |
D |
D |
D |
D |
D |
|
rs2233319 |
M67V |
1 |
D |
D |
D |
D |
D |
D |
D |
|
rs201348291 |
P298S |
1 |
D |
D |
D |
D |
D |
D |
D |
|
rs368404338 |
R322C |
1 |
D |
D |
D |
D |
D |
D |
D |
|
rs144714216 |
R343H |
1 |
D |
D |
D |
D |
D |
D |
D |
|
rs368584363 |
Y7H |
2 |
D |
D |
D |
D |
D |
D |
D |
|
rs373637595 |
A221T |
2 |
D |
D |
D |
D |
D |
D |
D |
|
rs201348291 |
P232S |
2 |
D |
D |
D |
D |
D |
D |
D |
|
rs144714216 |
R277H |
2 |
D |
D |
D |
D |
D |
D |
D |
|
rs201759485 |
V189A |
3 |
D |
D |
D |
D |
D |
D |
D |
|
rs201348291 |
P217S |
3 |
D |
D |
D |
D |
D |
D |
D |
|
rs144714216 |
R262H |
3 |
D |
D |
D |
D |
D |
D |
D |
|
rs146613168 |
R262C |
3 |
D |
D |
D |
D |
D |
D |
D |
3.3 Analysis of Protein-Protein Interaction:
The STRING tool (available at: https://string-db.org/cgi/network?taskId=b022 LqlKmSV8&sessionId=brixGRS2HYsO), provided results represented in figure 1. The server indicated that NDRG1 interacts with 11 proteins, including p53 cell tumor antigen (TP53), N-myc proto-oncogene protein (MYCN), serine/threonine-protein kinase (SGK1), NDRG2 protein (NDRG2), Myc proto-oncogene protein (MYC), WNT1-inducible signaling pathway protein 1 (WISP1), RAC -alpha serine/threonine-protein kinase (AKT1), and cadherin 1 (CDH1).
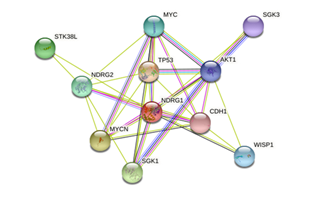
Figure1:Resistin protein-protein interaction network using a STRING server (https://string-db.org/cgi/network?taskId=b022 LqlKmSV8&sessionId=brixGRS2HYsO). Colored nodes: query proteins and first layer of interactors; white nodes: second layer of interactors; empty nodes: proteins of unknown 3D structure; filled nodes: some 3D structure is known or predicted.
4. Discussion
The NDRG1 gene encodes a protein present in the cytosol and expressed in epithelial cells. While there is evidence that its activation has anti-metastatic action, other studies suggest that the protein expression is related to less tumor differentiation, more aggressive metastatic potential and, therefore, worse prognosis (10, 11). In fact, NDRG1 gene expression in breast (21, 28), liver (21, 22, 23, 25), lung (26) and cervical cancer (24) was positively correlated with disease recurrence and considered an indicator of poor prognosis for patient survival (26). On the other hand, the expression of NDRG1 in prostate (30), colon (29) and esophagus tumors (28) was associated with a favorable clinical evolution.
An interesting strategy to investigate the role of a specific gene in the pathophysiology of a disease is the evaluation of the impact of this gene´s polymorphisms on the risk to develop this disease and/or its outcomes. Unfortunately, more than 300 missense polymorphisms have been reported in the human NDRG1 gene, and that number is still increasing. Not all of these polymorphisms include amino acid changes, and not all changes significantly affect the structure or function of the corresponding protein.
Data on the influence that nsSNPs of the NDRG1 gene have in the development of thyroid diseases, and their association with thyroid nodule clinical presentation and outcome have not yet been described in the literature. In order to select those that have the greatest potential to cause deleterious effects and influence the aetiopathogenesis of thyroid cancer, we undertook a thorough bioinformatics analysis of NDRG1 SNPs. We demonstrated that rs201348291 and rs151322132, two SNPs still poorly acknowledged, are potential markers of malignancy and may be related to the characteristics of thyroid cancer.
5. Conclusion
Our analysis indicated that these two nsSNPs deserve further validation, but our data have an obvious limitation, as we only performed in silico analyses. Clinical large-scale studies in different ethnic populations and laboratory experiments may provide more robust validation of our results. On the other hand, we provided solid foundation in the selection of SNPs of potential utility in the risk, evolution and prognosis of thyroid cancer patients.
6. Acknowledgments
The authors thank the group of the Laboratory of Cancer Molecular Genetics (GEMOCA) of the School of Medical Sciences (FCM-UNICAMP) for technical support. This study received financial support from the São Paulo Research Foundation (FAPESP grant 2020/16557-8) and INCT-TeraNano (CNPq nº 403193/2022-2). GBB received a research support from FAPESP. LSW is a Category 1 Research Fellow at the National Council for Scientific and Technological Development (CNPq).
7. Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
8. Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request
References
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome research 8 (1998): 1229–1231.
- Rajasekaran R, Sudandiradoss C, Doss CGP, et al. Identification and in silico analysis of functional SNPs of the BRCA1 gene. Genomics 90 (2007): 447–452.
- Elkhattabi L, Morjane I, Charoute H, et al. In Silico Analysis of Coding/Noncoding SNPs of Human RETN Gene and Characterization of Their Impact on Resistin Stability and Structure. Peterson JM, editor. Journal of Diabetes Research [Internet] 10 (2019).
- Dabhi B, Mistry KN. In silico analysis of single nucleotide polymorphism (SNP) in human TNF-α gene. Meta Gene 2 (2014): 586–595.
- AbdulAzeez S, Borgio JF. In-Silico Computing of the Most Deleterious nsSNPs in HBA1 Gene. PloS one 11 (2016)
- Ellen TP, Ke Q, Zhang P, et al. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis 29 (2008): 2–8.
- Sun J, Zhang D, Bae D-H, et al. Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 34 (2013): 1943–1954.
- Bandyopadhyay S, Pai SK, Gross SC, et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer research 63(8) (2003): 1731–1736.
- Hao X, Zhang S, Hu W, et al. Characterization of the prognostic values of the NDRG family in gastric cancer. Therapeutic Advances in Gastroenterology 12 (2019)
- Gerhard R, Nonogaki S, Fregnani JHTG, et al. NDRG1 protein overexpression in malignant thyroid neoplasms, Clinics. scielo 65 (2010): 757–762.
- Mosquera JM, Beltran H, Park K, et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia (New York, NY) 15 (2013): 1–10.
- Chua M-S, Sun H, Cheung ST, et al. Overexpression of NDRG1 is an indicator of poor prognosis in hepatocellular carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 20 ( 2007): 76–83.
- Cheng J, Xie H-Y, Xu X, et al. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer letters 310 ( 2011): 35–45.
- Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research 29 (2001): 308–311.
- Bendl J, Stourac J, Salanda O, et al. PredictSNP: Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations. PLOS Computational Biology [Internet] 10 (2014)
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research 31 (2003): 3812–3814.
- Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome research 15 ( 2005): 978–986.
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics (Oxford, England) 22 (2006): 2729–2734.
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic acids research 35 (2007): 3823–3835.
- Jr J. Using Stata with PHASE and Haploview: Commands for Importing and Exporting Data. Stata Journal 10(2010): 359–368.
- Mao Z, Sun J, Feng B, et al. The Metastasis Suppressor, N-myc Downregulated Gene 1 (NDRG1), Is a Prognostic Biomarker for Human Colorectal Cancer. PLoS ONE 8 ( 2013): 68206.
- Zhang X, Feng B, Zhu F, et al. N-myc downstream-regulated gene 1 promotes apoptosis in colorectal cancer via up-regulating death receptor 4. Oncotarget 8 (2017): 82593–82608.
- Cheng J, Xie HY, Xu X, et al. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett 310 (2011): 35–45.
- Nishio S, Ushijima K, Tsuda N, et al. Cap43/ NDRG1/Drg-1 is a molecular target for angiogenesis and a prognostic indicator in cervical adenocarcinoma. Cancer Lett 264 (2008): 36–43.
- Akiba J, Ogasawara S, Kawahara A, et al. N-myc downstream regulated gene 1 (NDRG1)/Cap43 enhances portal vein invasion and intrahepatic metastasis in human hepatocellular carcinoma. Oncol Rep 20 (2008): 1329–1335.
- Azuma K, Kawahara A, Hattori S, et al. NDRG1/ Cap43/Drg-1 may predict tumor angiogenesis and poor outcome in patients with lung cancer. J Thorac Oncol 7 (2012): 779–789.
- Nagai MA, Gerhard R, Fregnani JH, et al. Prognostic value of NDRG1 and SPARC protein expression in breast cancer patients. Breast Cancer Res Treat 126 (2011): 1–14.
- Ando T, Ishiguro H, Kimura M, et al. Decreased expression of NDRG1 is correlated with tumor progression and poor prognosis in patients with esophageal squamous cell carcinoma. Dis Esophagus 19 (2006): 454– 458.
- Strzelczyk B, Szulc A, Rzepko R, et al. Identification of high-risk stage II colorectal tumors by combined analysis of the NDRG1 gene expression and the depth of tumor invasion. Ann Surg Oncol 16 (2009): 1287–1294.
- Song Y, Oda Y, Hori M, et al. N-myc downstream regulated gene-1/Cap43 may play an important role in malignant progression of prostate cancer, in its close association with E-cadherin. Hum Pathol 41 (2010): 214– 222.
- Chun-Chi Chang, Ya-Sian Chang, Hsi-Yuan Huang, et al. Determination of the mutational landscape in Taiwanese patients with papillary thyroid cancer by whole-exome sequencing Yhupa 78 (2018):151-158.
- Ai R, Sun Y, Guo Z, Wei W, et al. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer biology & therapy 17 (2016): 943-954.
- Chang CC, Chang YS, Huang HY, et al. Determination of the mutational landscape in Taiwanese patients with papillary thyroid cancer by whole-exome sequencing. Hum Pathol 78 (2018):151-158.
