Altered Uterine Gene Expression in Lean and Obese Mice Following Maternal Oxytocin
Article Information
Shelly Soni1, Prodyot K Chatterjee2, Frances F Hsieh1, Xiangying Xue2, Nina Kohn3, Swati Madankumar2, Burton Rochelson1 and Christine N Metz1,2*
1Department of Obstetrics and Gynecology, Donald and Barbara Zucker School of Medicine at Hofstra/Northwell, 500 Hofstra Blvd, Hempstead, NY, USA
2Feinstein Institutes for Medical Research at Northwell Health, Institute of Molecular Medicine, 350 Community Dr, Manhasset, NY, USA
3The Biostatistics Unit, Feinstein Institutes for Medical Research at Northwell Health, 350 Community Dr, Manhasset, NY, USA
*Corresponding author: Christine N Metz, The Feinstein Institutes for Medical Research, 350 Community Drive, Manhasset, NY 11030 USA.
Received: 09 August 2022; Accepted: 16 August 2022; Published: 07 September 2022
Citation: Shelly Soni, Prodyot K Chatterjee, Frances F Hsieh, Xiangying Xue, Nina Kohn, Swati Madankumar, Burton Rochelson and Christine N Metz. Altered Uterine Gene Expression in Lean and Obese Mice Following Maternal Oxytocin. Journal of Women’s Health and Development 5 (2022): 206-220.
View / Download Pdf Share at FacebookAbstract
Background: Obese women exhibit higher rates of failed labor inductions with oxytocin. To investigate the mechanisms underlying parturition dysfunction in obese populations, we examined the changes in uterine gene expression profiles in lean and obese mice at term, with and without maternal oxytocin administration.
Methods: Female C57BL/6 mice were fed either a high-fat or regular-lean diet for 6 weeks prior to conception and throughout pregnancy. At term, dams were given saline or oxytocin, with a second group of obese mice receiving high-dose oxytocin. Six hours later, uterine gene expression for 30 select transcripts associated with parturition (e.g. gap junctions, relaxation/contractility pathways, and oxytocin signaling) and obesity were analyzed by quantitative real time PCR.
Results: Lean and obese uteri, at baseline, showed differential gene expression patterns at term. Oxytocin significantly altered the expression of numerous myometrial transcripts associated with parturition (gap junctions, relaxation/contractility pathways, and oxytocin signaling). The expression of numerous oxytocin-responsive genes depended on the dams’ body masses (lean vs. obese), with either blunted effects or no effects of oxytocin observed in obese mice vs. lean mice. Additionally, high-dose oxytocin did not consistently regulate parturition-related gene expression in obese uteri. In summary, gene expression patterns significantly differed in lean vs. obese uteri at term in the presence and absence of maternal oxytocin. Lean uteri were more responsive to oxytocin than obese uteri, even at higher doses of oxytocin.
Conclusions: These findings support that blunted oxytocin responsiveness in obese uteri may contribute to obesity-related labor dysfunction.
Keywords
Labor Induction; Obesity; Oxytocin; Parturition; Uterine Contraction
Labor Induction articles Labor Induction Research articles Labor Induction review articles Labor Induction PubMed articles Labor Induction PubMed Central articles Labor Induction 2023 articles Labor Induction 2024 articles Labor Induction Scopus articles Labor Induction impact factor journals Labor Induction Scopus journals Labor Induction PubMed journals Labor Induction medical journals Labor Induction free journals Labor Induction best journals Labor Induction top journals Labor Induction free medical journals Labor Induction famous journals Labor Induction Google Scholar indexed journals Obesity articles Obesity Research articles Obesity review articles Obesity PubMed articles Obesity PubMed Central articles Obesity 2023 articles Obesity 2024 articles Obesity Scopus articles Obesity impact factor journals Obesity Scopus journals Obesity PubMed journals Obesity medical journals Obesity free journals Obesity best journals Obesity top journals Obesity free medical journals Obesity famous journals Obesity Google Scholar indexed journals Oxytocin articles Oxytocin Research articles Oxytocin review articles Oxytocin PubMed articles Oxytocin PubMed Central articles Oxytocin 2023 articles Oxytocin 2024 articles Oxytocin Scopus articles Oxytocin impact factor journals Oxytocin Scopus journals Oxytocin PubMed journals Oxytocin medical journals Oxytocin free journals Oxytocin best journals Oxytocin top journals Oxytocin free medical journals Oxytocin famous journals Oxytocin Google Scholar indexed journals Parturition articles Parturition Research articles Parturition review articles Parturition PubMed articles Parturition PubMed Central articles Parturition 2023 articles Parturition 2024 articles Parturition Scopus articles Parturition impact factor journals Parturition Scopus journals Parturition PubMed journals Parturition medical journals Parturition free journals Parturition best journals Parturition top journals Parturition free medical journals Parturition famous journals Parturition Google Scholar indexed journals Uterine Contraction articles Uterine Contraction Research articles Uterine Contraction review articles Uterine Contraction PubMed articles Uterine Contraction PubMed Central articles Uterine Contraction 2023 articles Uterine Contraction 2024 articles Uterine Contraction Scopus articles Uterine Contraction impact factor journals Uterine Contraction Scopus journals Uterine Contraction PubMed journals Uterine Contraction medical journals Uterine Contraction free journals Uterine Contraction best journals Uterine Contraction top journals Uterine Contraction free medical journals Uterine Contraction famous journals Uterine Contraction Google Scholar indexed journals neuropeptide articles neuropeptide Research articles neuropeptide review articles neuropeptide PubMed articles neuropeptide PubMed Central articles neuropeptide 2023 articles neuropeptide 2024 articles neuropeptide Scopus articles neuropeptide impact factor journals neuropeptide Scopus journals neuropeptide PubMed journals neuropeptide medical journals neuropeptide free journals neuropeptide best journals neuropeptide top journals neuropeptide free medical journals neuropeptide famous journals neuropeptide Google Scholar indexed journals exogenously articles exogenously Research articles exogenously review articles exogenously PubMed articles exogenously PubMed Central articles exogenously 2023 articles exogenously 2024 articles exogenously Scopus articles exogenously impact factor journals exogenously Scopus journals exogenously PubMed journals exogenously medical journals exogenously free journals exogenously best journals exogenously top journals exogenously free medical journals exogenously famous journals exogenously Google Scholar indexed journals physiological articles physiological Research articles physiological review articles physiological PubMed articles physiological PubMed Central articles physiological 2023 articles physiological 2024 articles physiological Scopus articles physiological impact factor journals physiological Scopus journals physiological PubMed journals physiological medical journals physiological free journals physiological best journals physiological top journals physiological free medical journals physiological famous journals physiological Google Scholar indexed journals cytoskeletal articles cytoskeletal Research articles cytoskeletal review articles cytoskeletal PubMed articles cytoskeletal PubMed Central articles cytoskeletal 2023 articles cytoskeletal 2024 articles cytoskeletal Scopus articles cytoskeletal impact factor journals cytoskeletal Scopus journals cytoskeletal PubMed journals cytoskeletal medical journals cytoskeletal free journals cytoskeletal best journals cytoskeletal top journals cytoskeletal free medical journals cytoskeletal famous journals cytoskeletal Google Scholar indexed journals calcium articles calcium Research articles calcium review articles calcium PubMed articles calcium PubMed Central articles calcium 2023 articles calcium 2024 articles calcium Scopus articles calcium impact factor journals calcium Scopus journals calcium PubMed journals calcium medical journals calcium free journals calcium best journals calcium top journals calcium free medical journals calcium famous journals calcium Google Scholar indexed journals
Article Details
1. Introduction
Labor is a complex physiological process requiring changes in the structure and function of the uterus and cervix. The transition from uterine quiescence to the onset of contractions results from increased levels of contractile stimulators and loss of uterine relaxation [1]. Phasic and coordinated contractions are pertinent for successful labor outcomes. Despite multiple studies attempting to understand this process, the precise mechanisms of labor onset are poorly understood. In the US, approximately 30% of women undergo labor induction [2]. In clinical practice, oxytocin (OXT), a neuropeptide, is exogenously administered to induce and augment labor [3, 4]. OXT binds to OXT receptors (OXTRs) to initiate a cascade of events resulting in gene and protein expression changes, as well as calcium release leading to cytoskeletal changes that culminate in myocyte contractions [5].
Obese patients experience delayed labor initiation, protracted courses of labor and higher rates of failed inductions [6-10]. In addition, obese women may require higher doses of OXT and longer durations of OXT for labor augmentation [4, 11-13]. These complexities may lead to more cesarean deliveries and increased maternal and neonatal morbidities and mortality among obese women [7, 10, 14-16]. The biologic mechanisms responsible for parturition dysfunction and reduced OXT responsiveness among obese women are not completely understood, creating an unmet need for improving the labor process for this ‘at-risk’ group. To our knowledge and based on a search of the literature, no prior study examined a panel of genes associated with parturition (including gap junctions and uterine relaxation and contraction pathways) in lean and obese uteri at term, with and without maternal OXT administration.
Based on the influence of obesity on parturition and OXT responsiveness, we hypothesized that maternal obesity (vs. normal weight) would alter gene expression patterns in the myometrium at baseline and following maternal OXT treatment. To test this hypothesis, we examined the effect of maternal obesity on myometrial gene expression and responsiveness to oxytocin (vs. saline) using targeted quantitative real time PCR (qPCR), focusing on gene expression associated with parturition (gap junctions, uterine contractility, and relaxation, and OXT signaling) and obesity. In addition, to mimic higher doses of OXT administered to obese women, we also examined the effect of high dose vs. regular dose OXT on uterine gene expression in obese mice.
2. Methods
2.1. Ethics Approval
The Institutional Animal Care and Use Committee (IACUC) of the Feinstein Institutes for Medical Research reviewed and approved the animal studies (IACUC #2015-053) prior to animal experimentation. All animal experimentation was in accordance with the guidelines for animal care provided by the National Institutes of Health. No human experimentation was performed.
2.2. Mouse Model of Pregnancy
After acclimation to normal environmental conditions with 12hrs light and dark cycles, female Mus musculus (mice, C57BL/6, 4-5 weeks old, from Charles River) were fed either high-fat diet (HFD, D-12492, Research Diets Inc) or standard regular chow diet (D12450B, Research Diets Inc). The HFD consisted of 60% of Kcal as fat and 20% Kcal as protein; the regular diet consisted of 10% of Kcal as fat and 20% of Kcal as protein. After 6 weeks on their respective diets, females (10-11 weeks old) were mated with regular diet-fed C57BL/6 males (10-15 weeks old, Charles River). Timed matings were set up in the late afternoons approximately twice weekly during proestrus (based on the assessment of external female genitalia and microscopic evaluation of vaginal cell morphology following lavage [17], using male-urine synchronized cycles). The following morning (before 7am) males were removed and females were checked for sperm plugs or the presence of sperm in vaginal lavage and for vaginal cell morphology (determined as the first day of gestation; gestational day (GD) 0.5). Females were weighed twice weekly to confirm timed pregnancies. Pregnant mice were maintained on their respective diets throughout the gestation. This model of gestational obesity using pre- and post-conception administration of the HFD is similar that previously described by Chang et al using C57BL/6 mice [18]; this HFD model in female C57BL/6 mice was chosen because it induces obesity, lipid derangements and hypercholesterolemia [19], mimicking obesity with lipid dysfunction.
On GD18.5 (at term, the mean gestation length for C57BL/6 mice is 19 days), mice (n=6-7 per group) were administered either saline (100μl) or 1U oxytocin (OXT, JHP Pharmaceuticals, Inc) administered in 100μl subcutaneously (s.c.) every 30 min for 2 hours (cumulative dose of 5U/mouse (equivalent to 0.334mg/kg), regular [reg]-dose). This dosing method was chosen based on veterinary OXT use in mice [20] and to avoid invasive pump implantation in term-pregnant mice. This dose was based on [21] and our prior studies showing that 5U OXT per day (n=7 dams/group) did not induce labor when compared to saline (all mice delivered within 15-20 hours of initial treatment [i.e. the expected time-frame]) and to avoid the delay in delivery observed with low dose maternal OXT [21]. On GD18.5 (at term), another set of obese dams (n=6) were given double the dose of OXT (2U/100μl s.c. every 30 min for 2 hours (cumulative dose of 10U/mouse, high [hi]-dose OXT)) to mimic a higher dose given to obese women. Six hours after the first OXT dose, dams were euthanized by CO2 asphyxiation and exsanguination; pups were delivered by C-section and uteri were collected. Note: none of the dams delivered within 6 hours of saline or OXT administration. Myometrial tissue was prepared as previously described [22]. Briefly, both uterine horns were dissected, the uterine tissue closest to the cervix was removed, opened and rinsed with PBS. Placental tissue was removed from the uterine tissue and care was taken to avoid the inclusion of decidua basalis. Myometrial tissue was immediately frozen in liquid nitrogen and then stored at -80°C for RNA isolation.
2.3. RNA Isolation and Gene Expression Studies
RNA was isolated using the RNeasy Universal Plus Mini Kit (Qiagen), which included a genomic DNA elimination step. RNA quality was analyzed using the Nanodrop spectrophotometer and the Bioanalyzer (Agilent Technologies Genomics). Total RNA samples with OD260:280 and OD260:230 ratios >1.9 and RIN values > 9.3 were used for analyses.
2.4. Experiment 1. Assessment of Uterine Contractility/Relaxation Pathways in Lean vs. Obese Dams ± OXT
A panel of 30 mRNA transcripts (Supplementary Table 1, see section of supplementary data given at the end of this article) was selected to assess obesity, gap junction-related pathways, and parturition-related pathways (contractility and relaxation, OXT signaling), as previously described in the literature. Targeted qPCR was performed using 6-7 uteri/group (lean±vehicle or OXT and obese±vehicle or OXT). Briefly, genomic DNA-free RNA (1μg per uterus) was reverse transcribed to double stranded cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with the following temperature cycles: 25°C for 10 min, 37°C for 120 min and heat inactivation at 85°C for 5 min. Real time PCR/qPCR using synthesized cDNA, specific primers and the Roche Universal Probe Library was performed using the Eurogentec qPCR MasterMix Plus (AnaSpec, Inc) (see Supplementary Table 1) and the Roche LightCycler 480 (each transcript was run in triplicate) using the following cycling programs: 50°C for 2 min and 95°C for 10 min for 1 cycle, followed by 45 cycles of 95°C for 15 sec and 60°C for 75 sec. Data were analyzed for relative changes in gene expression (using Roche LightCycler software and Microsoft Excel) and reported as fold-changes by the 2-ΔΔCt method using Gapdh for normalizing transcript levels. Note: Gapdh transcripts were shown to be stable across mice, conditions, and experimental days.
2.5. Experiment 2. Assessment of OXT Dosing in Obese Dams
The effect of high (hi)-dose oxytocin administration to obese dams (n=6-7 per group; obese+vehicle, obese+reg-dose OXT, and obese hi-dose OXT) on the expression of 30 specific genes implicated in parturition-related pathways (contractility and relaxation, OXT signaling) and obesity was analyzed by qPCR, as described above.
2.6. Statistical Analysis
2.6.1. Sample sizes: Sample sizes for the 2x2 factorial design (experiment 1) and the dose escalation study (experiment 2) were based on published studies using OXT for studying labor induction and uterine contractility in mice [21, 23].
2.6.2. Experiment 1. Assessment of Uterine Contractility/Relaxation Pathways in Lean vs. Obese Dams ± OXT: Based on the study design two-way analysis of variance (ANOVA) was used to examine the association between expression levels and group (lean, obese) and OXT (no, yes) (2x2 factorial design). The interaction between group (lean vs. obese) and treatment (no OXT, OXT) was included in the model. If the interaction was significant, then the following pre-specified comparisons were examined within the ANOVA model: lean mice ± OXT; obese mice ± OXT; and lean vs. obese mice (–OXT). For these comparisons, a Bonferroni adjustment was used, such that p<0.0167 was considered significant. If the interaction between group and OXT was not significant, it was removed and the main effects of group (lean, obese) and treatment (no OXT, OXT) were examined; p<0.05 was considered significant. It should be noted that the purpose of the interaction term is to examine whether the change in expression levels with treatment (no OXT, OXT) differed depending on group (lean/control, obese). In those models where the interaction was not significant, the interaction term was removed and only the effects of treatment and group on expression levels were examined.
2.6.3. Experiment 2. Assessment of OXT Dosing in Obese Dams: Based on the study design analysis of variance, (ANOVA) was used to examine the association between expression levels and OXT (no, OXT, regular dose OXT and high dose OXT; n=6/group). If there was a significant effect of OXT, pairwise comparisons were carried out within the ANOVA model. A Bonferroni adjustment was used, such that p<0.0167 was considered significant for these comparisons.
For experiments 1 and 2, the log2 transformation was used to better meet the assumptions of the ANOVA model. Log2 transformed mRNA expression data more closely follows a normal distribution than untransformed expression data. For all genes in experiments 1 and 2 vehicle-treated lean mice (who did not receive OXT) were the reference group (i.e. all levels were divided by the mean expression level in this group). Summary statistics are given as least squares means and their associated 95% confidence intervals determined from the ANOVA model and then transformed back to non-log scale expression levels (fold-change in expression).
3. Results
3.1 Maternal weight gain and litter assessments
The average weights of lean and obese dams at term were statistically different, with obese mice weighing approximately 33% more (44.1±5.7g [obese] vs. 33±3.6g [lean], p<0.0001). Also, pup weights differed, with heavier pups in the obese group when compared to the lean group (0.98±0.2g [obese] vs. 0.87±0.1g [lean], p=0.0005). Litter sizes were comparable in lean and obese groups (7.5±1.7 vs. 8.4±2.5, p=0.3).
3.2 Differential Expression of Uterine Contractility/Relaxation Pathways in Lean vs. Obese Dams following Maternal OXT Administration
Using a 2x2 study design in lean and obese mice, we examined the effects of maternal OXT administration (or vehicle) on a panel of genes associated with OXT signaling, parturition pathways (e.g. uterine contractility and relaxation pathways), and obesity (See Supplementary Table 1 for the full list of genes). Among the 30 genes analyzed, 11 genes showed significant interactions – i.e. the effect of OXT on gene expression depended on whether the animals were lean or obese (Table 1 and Supplementary Table 2 for full data set). These genes included Edn1, Gucy1a3, Gucy1b3, Kcnq1, Mapk1, Mylk, Mypt3, Npr1, Pgrmc1, Pgrmc2, and Prkca (Table 1). In lean mice, the uterine expression of all 11 genes was significantly reduced by maternal OXT administration, whereas only Mypt3 and Npr1 expression was significantly reduced in obese uteri. However, greater effects were observed in lean mice (Table 1). Finally, the expression of Edn1, Kcnq1, Npr1, and Prkca was significantly higher in lean vs. obese uteri, irrespective of OXT treatment (Table 1).
For the remaining genes, where no significant interactions were observed, the main effects of group (lean vs. obese) and treatment (saline vs. OXT) were analyzed. Among these 19 genes, 15 showed significant differences (p<0.05) (Table 2). Almost half of these genes were differentially expressed in lean vs. obese uteri irrespective of maternal OXT administration, with higher expression in lean vs. obese uteri for Avpr, Dbn1, Oxtr, Pdgfra, Prkcg, and Ptgs1 or lower expression in lean vs. obese uteri for Lep, Nampt, and Oxt (Table 2). More than half of the genes examined showed significant responsiveness to maternal OXT treatment irrespective of dam weights, including Adipoq, Avp, Dbn1, Ghrl, Nampt, Nppa, Oxtr, Plcb1, Prkacb, Prkcg, and Ptgs1 (Table 2). Specifically, Avp, Dbn1, Ghrl, Oxtr, Plcb1, and Prkcg were increased following maternal OXT administration, while Adipoq, Nampt, Nppa, Prkacb, and Ptgs1 were decreased (Table 2).
|
Gene |
Lean |
Obese |
|||||
|
-OXT |
+OXT |
-OXT |
+OXT |
||||
|
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
p-value for interactiona |
Main Effect Comparisonsb |
p-value |
|
|
Edn1
|
0.992 (0.882, 1.115) |
0.575 (0.516, 0.641) |
0.599 (0.537, 0.667) |
0.516 (0.463, 0.575) |
0.0012 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0569 <.0001 |
|
Gucy1a3
|
0.990 (0.852, 1.151) |
0.493 (0.429, 0.566) |
0.769 (0.669, 0.884) |
0.633 (0.551, 0.728) |
0.0013 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0523 0.0180 |
|
Gucy1b3
|
0.990 (0.861, 1.139) |
0.522 (0.459, 0.595) |
0.781 (0.686, 0.890) |
0.661 (0.581, 0.753) |
0.0012 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0725 0.0173 |
|
Kcnq1
|
0.986 (0.817, 1.190) |
0.622 (0.522, 0.740) |
0.581 (0.488, 0.692) |
0.538 (0.452, 0.641) |
0.0356 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0011 0.5257 0.0003 |
|
Mapk1
|
0.998 (0.912, 1.092) |
0.712 (0.655, 0.774) |
0.822 (0.756, 0.893) |
0.878 (0.808, 0.955) |
<.0001 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.2569 0.0033 |
|
Mylk |
0.952 (0.730, 1.243) |
0.395 (0.309, 0.506) |
0.582 (0.455, 0.745) |
0.465 (0.363, 0.595) |
0.0130 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.1958 0.0100 |
|
Mypt3
|
0.997 (0.897, 1.108) |
0.533 (0.483, 0.588) |
0.743 (0.674, 0.820) |
0.547 (0.496, 0.603) |
0.0031 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0001 0.0003 |
|
Npr1 |
0.994 (0.826, 1.198) |
0.462 (0.389, 0.548) |
0.629 (0.530, 0.748) |
0.435 (0.366, 0.517) |
0.0281 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0047 0.0011 |
|
Pgrmc1 |
0.994 (0.872, 1.132) |
0.720 (0.638, 0.812) |
0.868 (0.769, 0.980) |
0.855 (0.758, 0.965) |
0.0169 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0011 0.8576 0.1296 |
|
Pgrmc2
|
0.985 (0.792, 1.224) |
0.667 (0.545, 0.816) |
0.909 (0.743, 1.112) |
1.079 (0.882, 1.320) |
0.0097 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0124 0.2260 0.5820 |
|
Prkca |
0.988 (0.856, 1.140) |
0.671 (0.587, 0.766) |
0.736 (0.644, 0.840 |
0.684 (0.599, 0.782) |
0.0251 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0005 0.4358 0.0049 |
|
a p<0.05 is significant b For these pre-specified comparisons, a Bonferroni adjustment was used such that p<0.0167 was considered significant. |
|||||||
Table 1: Gene Expression Comparing Groups of Mice (lean vs. obese) and Treatment (± OXT) With Significant Interactions.
|
Gene |
Lean |
Obese |
||||
|
-OXT |
+OXT |
-OXT |
+OXT |
Main Effect |
||
|
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
Comparisonsa |
p-value |
|
|
Adipoq |
0.556 (0.209, 1.484) |
0.167 (0.067, 0.414) |
0.683 (0.275, 1.693) |
0.203 (0.082, 0.504) |
Obese vs Lean No Oxt vs Oxt |
0.6515 0.0110 |
|
Avp |
0.706 (0.296, 1.687) |
1.958 (0.874, 4.385) |
1.420 (0.594, 3.393) |
2.707 (1.208, 6.062) |
Obese vs Lean No Oxt vs Oxt |
0.2228 0.0476 |
|
Avpr
|
0.941 (0.706, 1.253) |
1.156 (0.887, 1.507) |
0.688 (0.527, 0.896) |
0.627 (0.481, 0.817) |
Obese vs Lean No Oxt vs Oxt |
0.0016 0.7015 |
|
Dbn1 |
0.994 (0.872, 1.133) |
1.418 (1.256, 1.602) |
0.838 (0.742, 0.946) |
1.145 (1.014, 1.293) |
Obese vs Lean No Oxt vs Oxt |
0.0031 <.0001 |
|
Ghrl |
0.982 (0.804, 1.200) |
1.545 (1.283, 1.861) |
0.846 (0.703, 1.019) |
1.268 (1.053, 1.527 |
Obese vs Lean No Oxt vs Oxt |
0.0638 <.0001 |
|
Lep |
0.571 (0.218, 1.493) |
0.396 (0.163, 0.965) |
1.513 (0.621, 3.687) |
1.096 (0.419, 2.867) |
Obese vs Lean No Oxt vs Oxt |
0.0323 0.4391 |
|
Nampt |
0.989 (0.763, 1.283) |
0.676 (0.531, 0.860) |
1.283 (1.009, 1.632) |
0.865 (0.680, 1.100) |
Obese vs Lean No Oxt vs Oxt |
0.0392 0.0027 |
|
Nppa |
0.994 (0.860, 1.150) |
0.797 (0.697, 0.912) |
0.890 (0.778, 1.019) |
0.717 (0.627, 0.821) |
Obese vs Lean No Oxt vs Oxt |
0.1094 0.0026 |
|
Oxt |
0.934 (0.512, 1.703) |
1.551 (0.889, 2.705) |
2.373 (1.361, 4.138) |
2.164 (1.241, 3.774) |
Obese vs Lean No Oxt vs Oxt |
0.0335 0.4840 |
|
Oxtr |
0.909 (0.683, 1.210) |
2.994 (2.299, 3.900) |
0.570 (0.438, 0.743) |
1.254 (0.963, 1.634) |
Obese vs Lean No Oxt vs Oxt |
<.0001 <.0001 |
|
Pdgfra |
0.986 (0.860, 1.131) |
0.956 (0.842, 1.085) |
0.867 (0.764, 0.984) |
0.792 (0.698, 0.899) |
Obese vs Lean No Oxt vs Oxt |
0.0157 0.3245 |
|
Plcb1 |
0.975 (0.816, 1.165) |
1.645 (1.395, 1.940) |
0.798 (0.677, 0.941) |
1.516 (1.285, 1.788) |
Obese vs Lean No Oxt vs Oxt |
0.0980 <.0001 |
|
Prkcab |
0.990 (0.891, 1.100) |
0.716 (0.650, 0.790) |
0.811 (0.736, 0.894) |
0.707 (0.642, 0.780) |
Obese vs Lean No Oxt vs Oxt |
0.0553 0.0002 |
|
Prkcg |
0.994 (0.810, 1.219) |
1.152 (0.954, 1.392) |
0.615 (0.509, 0.743) |
0.783 (0.648, 0.946) |
Obese vs Lean No Oxt vs Oxt |
<.0001 0.0426 |
|
Ptgs1 |
0.989 (0.829, 1.181) |
0.587 (0.499, 0.692) |
0.670 (0.569, 0.789) |
0.539 (0.457, 0.635) |
Obese vs Lean No Oxt vs Oxt |
0.0116 0.0003 |
|
aFor these transcripts the interaction between group/BMI and OXT treatment (yes,no) was not significant (p>0.05) so the interaction was removed and the main effects of group and treatment were examined. Therefore, p<0.05 was considered significant. |
||||||
Table 2: Gene Expression Comparing Groups of Mice (lean vs. obese) and Treatment (± OXT) With Significant Interactions.
3.3. OXT Dosing in Obese Dams Results in Blunted Uterine Gene Expression Responses
Next, we examined the differences in the expression of uterine genes following vehicle (no OXT) vs. reg-dose OXT vs. hi-dose OXT administered to obese dams at term. The full dataset is in Supplementary Table 3. Among the same 30 genes analyzed in experiment 1, this dose escalation study revealed that 9 genes (Dbn1, Ghrl, Gucy1a3, Mypt3, Npr1, Oxtr, Plcb1, Prkcg, and Ptgs2) showed an overall significant dose effect (vs. vehicle) (Figure 1). Although Npr1 showed an overall significant p value (p<0.05) across all doses (vehicle, reg-dose, and hi-dose), no pairwise comparisons were significant (Supplementary Table 3) and thus it was not included in Figure 1. Among the 8 remaining genes, all increased following maternal OXT, except for Mypt3 and Gucy1a3, which decreased with OXT (Figure 1). While 7 genes showed significant differential expression for pairwise comparisons of hi-dose OXT vs. no OXT (vehicle) in obese mice, most exhibited a ‘plateau effect’ with increasing OXT doses in obese mice (i.e. a ‘blunted OXT response’) (Figure 1). Only Ptgs2 showed a significant difference for the most clinically relevant comparison, reg-dose OXT vs. hi-dose OXT (p=0.0083) (Figure 1).
4. Discussion
Rising rates of obesity among women of reproductive age are concerning [24] and there is ample evidence that obesity negatively impacts parturition, resulting in delayed labor, higher rates of labor dystocia, and poor outcomes [6-10, 14, 16]. The high prevalence of obesity may contribute to the high percentage of pregnant women receiving OXT to facilitate labor in the US [2]. While several baseline differences in lean vs. obese uteri/myometrium have been described [25] and some limited gene and protein expression profiling of the myometrium in pregnancy and in labor have been performed [26], the effects of obesity on myometrial maturation and labor induction and progression are incompletely understood.
Using a 2x2 study design to compare lean (±OXT) and obese (±OXT) groups of mice for gene expression involved in uterine contraction and relaxation, as well as labor and obesity (e.g. genes related to adipokines, gap junctions, and inflammation) allowed assessment of an interaction between obesity and OXT-relating signaling during parturition. Nine genes showed significant interactions, including Edn1, Gucy1a3, Gucy1b3, Kcnq1, Mapk1, Mylk, Pgrmc1, Pgrmc2, and Prkca, with OXT responsiveness only in lean mice (Table 1) – supporting that the OXT effect depended on body mass. Only Npr1 and Mypt3 (whose gene products play roles in the relaxation pathway) were significantly downregulated in both lean and obese mice by OXT, with greater effects in lean mice (Table 1). These data identify several myometrial mRNA transcripts implicated in reduced OXT responsiveness in obese mice (Figure 2).
For the remaining genes that showed no significant interactions (i.e. the effect of OXT did not depend on the dams’ body mass), the main effects of group (lean vs. obese) and treatment (no OXT vs. OXT) identified several genes implicated in parturition that were differentially expressed in obese and lean uteri (Table 2 and Figure 2). The expression of uterine contractility-related genes that encode OXTR (Oxtr) and phospholipase C beta (Pl cb1), a downstream effector of G-protein-coupled receptor signaling that has central roles in labor, were significantly higher in lean uteri and both genes were significantly upregulated in lean and obese dams following maternal OXT (p<0.0001). To our knowledge and based on a review of the literature, Plcb1 expression has not been assessed in lean vs. obese uteri. Interestingly, maternal obesity does not appear to influence myometrial OXTR gene or protein expression in humans [27]. This discrepancy may be due to differences in sampling sites, timing, or species. Clearly, assessment of myometrial OXTR protein expression and functional assessment of contractility ex vivo are warranted as hypercholesterolemia is associated with labor dysfunction and impaired OXT responsiveness [28, 29]. Cholesterol treatment of isolated uterine smooth muscle cells reduces contractility [30], possibly by reducing membrane fluidity and impairing OXTR function, as previously described for other membrane receptors [31].
Previous studies using a similar HFD model in rats reported decreased Cx43 (encoded by Gja1) protein expression in the pregnant uteri of obese vs. lean rats [32]. Cx43 is a major myometrial gap junction protein proposed to synchronize myometrial contractions [33]. In our study, Gja1 (encodes Cx43) was not differentially expressed in lean vs. obese mice (Supplementary Table 2). This difference may be related to the difference in species and/or mRNA vs. protein assessment. Interestingly, we found that Dbn1 (that encodes a Cx43 binding partner) was differentially expressed in lean and obese myometrium, with higher expression in lean myometrium (Table 2). While Dbn1 is upregulated in the pregnant myometrium [34], little is known about its function and to our knowledge its expression has not been linked to obesity.
A panel of adipokine genes was also examined based on their proposed contribution to labor dysfunction and reports that adipokines inhibit uterine contractility [35-39]. Originally thought to be expressed only by adipocytes, adipokine mRNAs and proteins are expressed by the myometrium (e.g. Lep/leptin [40, 41], Adipoq/adiponectin [42, 43], Nampt/visfatin [44], and Ghrl/ghrelin [45]). Not all studies report similar findings; these inconsistencies may be due to detection sensitivity, species differences, and/or tissue state (pregnant, non-pregnant, healthy, and pathologic). Consistent with obesity-induced leptin expression [46]. Lep was more highly expressed in obese uteri; however, it was unchanged by OXT (Supplementary Table 2). By contrast, OXT altered Adipoq, Ghrl and Nampt mRNA expression in both lean and obese uteri (Table 2). While these data suggest that OXT may not exert differential effects in obese uteri through these adipokine mRNAs, adipokine receptor expression should be examined.
Clinically, since obese patients may require higher OXT doses to augment labor [4, 11-13], we performed a dose escalation study using the same obesity model and gene panel comparing hi- vs. reg-dose OXT. Only one gene (among 30 examined), Ptgs2, showed a significant difference when comparing the clinically relevant comparison of hi- vs. reg-OXT doses (Figure 1, lower panel). Previous studies report that OXT upregulates Ptgs2 gene and COX-2 protein expression using cultured endometrial epithelial cells, supporting its role in mediating OXT-induced PGF2α production by these cells [47]. In addition, COX-2 expression is increased during labor [48]. The major differences between our study and prior studies were the use of an in vivo model and analysis of myometrial tissues vs. cultured epithelial cells. Interestingly, COX-2 inhibition has been proposed to mediate myometrial relaxation in vitro [49]. Overall, the results of the dose escalation study support that obesity reduces uterine OXT-responsiveness and suggest that future research focusing on enhancing OXTR responsiveness in the setting of obesity may be informative.
This study has several limitations. First, we only assessed uterine gene expression at one time point (6 hours post-OXT) and therefore, we lack a timeline of gene expression changes. Second, the reg-dose of OXT was relatively high; however, it was based on previous experimental work investigating the effects of OXT on parturition [21]. In our preliminary experiments, this dose did not induce labor and we avoided delayed labor reported with lower OXT doses OXT [21]. In addition, we did not assess global gene expression. Nor did we measure protein products or evaluate functional myometrial contractility. However, results from prior studies support that obesity/hypercholesterolemia (a feature of this obesity model) is associated with labor dysfunction, altered uterine contractility and OXT responsiveness [28, 29, 32] and together with our data, support future studies using a similar model. Finally, there are inherent limitations in translating findings from pregnant mice to pregnant humans.
5. Conclusions
In conclusion, lean and obese uteri exhibit significantly different gene expression profiles and obese uteri show blunted gene expression effects reflecting reduced baseline differences and OXT responsiveness (Figure 2). These effects may contribute to uncoordinated contractions and obesity-associated labor dystocia. For providing optimal delivery experiences with fewer adverse outcomes, future studies should focus on 1) defining body mass-based OXT dosing regimens through pre-clinical (animal) and clinical dosing studies and 2) investigating ways to improve OXT responsiveness and regulate uterine contractility and relaxation, using functional studies.
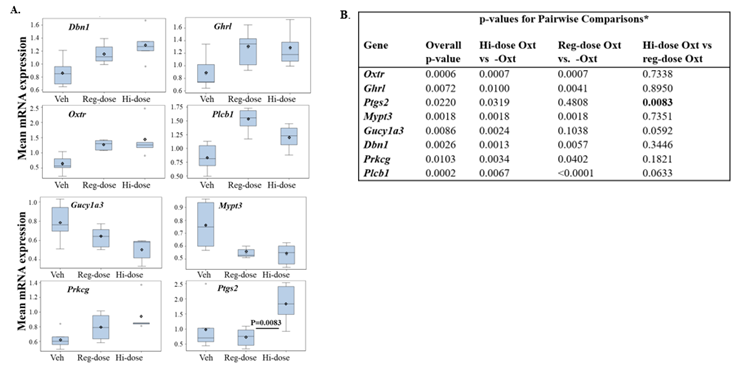
Figure 1: Variable Effects of Increasing OXT Doses on Uterine Gene Expression in Obese Dams.
Obese dams were treated with either vehicle (Veh), regular dose OXT (1U/100μl every 30min for 2h, reg-dose) or high dose (2U/100μl every 30min for 2h, hi-dose). (A) A significant interaction was found for Dbn1, Ghrl, Mypt3, Gucy1a3, Oxtr, Plcb1, Prkcg, and Ptgs2 genes, which were then analyzed using pairwise comparisons using ANOVA model. Box plots in the above panels describe the distribution of gene expression data in uteri from obese mice following treatment saline, reg-dose, or hi-dose OXT. ◊ = mean; center line = median; top and bottom of boxes = 75%-ile and 25%-ile, respectively; whiskers extend to minimum and maximum with outliers shown separately. (B) P values for the overall and pairwise comparisons are shown in the table; * for pairwise comparisons, a Bonferroni adjustment was used, such that p<0.0167 was considered significant.
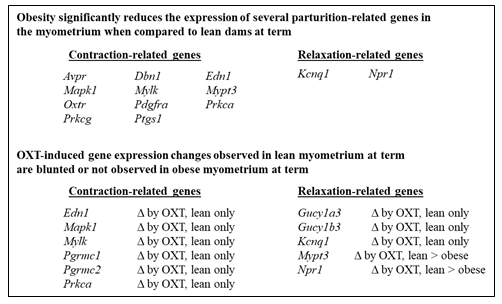
Figure 2: Differential Expression of Parturition-Related Myometrium Genes in Setting of Obesity.
Several contraction- and relaxation-related genes show differential expression in the myometrium of lean vs. obese pregnant mice, with significantly reduced gene expression in obese vs. lean mice (upper panel). In addition, several contraction- and relaxation-related genes in the myometrium exhibit decreased OXT responsiveness in obesity pregnant mice when compared to lean pregnant mice (lower panel). Δ = change.
Declarations
Ethics Approval and Consent to Participate/Consent for Publication
The Institutional Animal Care and Use Committee (IACUC) of the Feinstein Institutes for Medical Research reviewed and approved the animal studies (IACUC #2015-053) prior to animal experimentation. All animal experimentation was in accordance with the guidelines for animal care provided by the National Institutes of Health. No human experimentation was performed. No human subjects were involved in this research and therefore, there was no consent process.
Availability of Data and Materials
All data generated and analyzed during this study are included in this published manuscript (or included in the Supplemental Data).
Competing Interests, Conflict of Interest
All authors confirmed that all methods were performed in accordance with the relevant guidelines and regulations in research. The authors declare that they have no competing interests or conflicts of interest that could be perceived as prejudicing the impartiality of the research reported.
Authors' Contributions
SS, FFH, BR and CNM conceived experiments and designed the study. SS, FFH, XX, and CNM performed animal experimentation, sample collections and sample processing. SS, FFH and PKC performed RNA isolation and assessment of RNA quality. SS and PKC performed qPCR; SS, PKC, SM and CNM contributed to data interpretation and preparation of Tables and Figures. SS, SM, and CNM wrote the manuscript with SS and SM. NK performed all data analyses and reviewed the results with the authors. All authors reviewed, edited. and approved the final manuscript, Tables and Figures, as well as Supplementary data.
SS is currently at the Center for Fetal Diagnosis and Treatment, Children's Hospital of Philadelphia, 34th Street and Civic Center Boulevard, 5 Wood, Philadelphia, PA 19104 USA
FFH is currently at Stamford Hospital, One Hospital Plaza, Stamford, CT 06905 USA
Acknowledgement
The authors would like to thank the staff of the Center for Comparative Physiology for assisting with the animal care and maintenance.
Funding
This work was supported by the Feinstein Institutes for Medical Research (CNM) and the Lax Family Foundation (BR).
References
- Smith R. Parturition. N Engl J Med 356 (2007): 271-283.
- Martin JA, Hamilton BE, Osterman MJK, et al. Births: Final Data for 2019. Natl Vital Stat Rep 70 (2021): 1-51.
- Blanks AM, Thornton S. The role of oxytocin in parturition. BJOG 110 (2003): 46-51.
- Hill M, Reed KL, Cohen WR. Oxytocin utilization for labor induction in obese and lean women. J Perinat Med 43 (2015): 703-706.
- Vrachnis N, Malamas FM, Sifakis S, et al. The oxytocin-oxytocin receptor system and its antagonists as tocolytic agents. Int J Endocrinol 2011 (2011): 350546.
- Bogaerts A, Witters I, Van den Bergh BR, et al. Obesity in pregnancy: altered onset and progression of labour. Midwifery 29 (2013): 1303-1313.
- Jain D, Khuteta R, Chaturvedi V, et al. Effect of body mass index on pregnancy outcomes in nulliparous women delivering singleton babies: observational study. J Obstet Gynaecol India 62 (2012): 429-431.
- Arrowsmith S, Wray S, Quenby S. Maternal obesity and labour complications following induction of labour in prolonged pregnancy. BJOG 118 (2011): 578-588.
- Dammer U, Bogner R, Weiss C, et al. Influence of body mass index on induction of labor: A historical cohort study. J Obstet Gynaecol Res 44 (2018): 697-707.
- Ellis JA, Brown CM, Barger B, et al. Influence of Maternal Obesity on Labor Induction: A Systematic Review and Meta-Analysis. J Midwifery Womens Health 64 (2019): 55-67.
- Soni S, Chivan N, Cohen WR. Effect of maternal body mass index on oxytocin treatment for arrest of dilatation. J Perinat Med 41 (2013): 517-521.
- Roloff K, Peng S, Sanchez-Ramos L, et al. Cumulative oxytocin dose during induction of labor according to maternal body mass index Int J Gynaecol Obstet 131 (2015): 54-58.
- Maeder AB, Vonderheid SC, Park CG, et al. Titration of Intravenous Oxytocin Infusion for Postdates Induction of Labor Across Body Mass Index Groups. J Obstet Gynecol Neonatal Nurs 46 (2017): 494-507.
- Harper A. Reducing morbidity and mortality among pregnant obese. Best Pract Res Clin Obstet Gynaecol 29 (2015): 427-437.
- Pevzner L, Powers BL, Rayburn WF, et al. Effects of maternal obesity on duration and outcomes of prostaglandin cervical ripening and labor induction. Obstet Gynecol 114 (2009): 1315-1321.
- Baeten JM, Bukusi EA, Lambe M. Pregnancy complications and outcomes among overweight and obese nulliparous women. Am J Public Health 91 (2001): 436-440.
- Byers SL, Wiles MV, Dunn SL, et al. Mouse estrous cycle identification tool and images PLoS One 7 (2012): e35538.
- Chang E, Hafner H, Varghese M, et al. Programming effects of maternal and gestational obesity on offspring metabolism and metabolic inflammation. Sci Rep 9 (2019): 16027.
- Wang CY, Liao JK. A mouse model of diet-induced obesity and insulin resistance. Methods Mol Biol 821 (2012): 421-433.
- Narver HL. Oxytocin in the treatment of dystocia in mice. J Am Assoc Lab Anim Sci 51 (2012): 10-17.
- Imamura T, Luedke CE, Vogt SK, et al., Oxytocin modulates the onset of murine parturition by competing ovarian and uterine effects. Am J Physiol Regul Integr Comp Physiol 279 (2000): R1061-1067.
- Salomonis N, Cotte N, Zambon AC, et al., Identifying genetic networks underlying myometrial transition to labor. Genome Biol 6 (2005): R12.
- Kawamata M, Yoshida M, Sugimoto Y, et al., Infusion of oxytocin induces successful delivery in prostanoid FP-receptor-deficient mice. Mol Cell Endocrinol 283 (2008): 32-37.
- Deputy NP, Dub B, Sharma AJ. Prevalence and Trends in Prepregnancy Normal Weight - 48 States, New York City, and District of Columbia, 2011-2015. MMWR Morb Mortal Wkly Rep 66 (2018): 1402-1407.
- Carlson NS, Hernandez TL, Hurt KJ. Parturition dysfunction in obesity: time to target the pathobiology. Reprod Biol Endocrinol 13 (2015): 135.
- Breuiller-Fouche M, Germain G. Gene and protein expression in the myometrium in pregnancy and labor. Reproduction 131 (2006): 837-850.
- Gregory SG, Anthopolos R, Osgood CE, et al., Association of autism with induced or augmented childbirth in North Carolina Birth Record (1990-1998) and Education Research (1997-2007) databases. JAMA Pediatr 167 (2013): 959-966.
- Padol AR, Sukumaran SV, Sadam A, et al., Hypercholesterolemia impairs oxytocin-induced uterine contractility in late pregnant mouse. Reproduction 153 (2017): 565-576.
- Mouzat K, Prod'homme M, Volle DH, et al., Oxysterol nuclear receptor LXRbeta regulates cholesterol homeostasis and contractile function in mouse uterus. J Biol Chem 282 (2007): 4693-4701.
- Smith RD, Babiychuk EB, Noble K, et al., Increased cholesterol decreases uterine activity: functional effects of cholesterol alteration in pregnant rat myometrium. Am J Physiol Cell Physiol 288 (2005): C982-988.
- Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci. 57 (2000): 1577-1592.
- Muir R, Ballan J, Clifford B, et al., Modelling maternal obesity: the effects of a chronic high-fat, high-cholesterol diet on uterine expression of contractile-associated proteins and ex vivo contractile activity during labour in the rat. Clin Sci (Lond) 130 (2016): 183-192.
- Garfield RE, Sims S, Daniel EE. Gap junctions: their presence and necessity in myometrium during parturition. Science 198 (1977): 958-960.
- Rehman KS, Yin S, Mayhew BA, et al., Human myometrial adaptation to pregnancy: cDNA microarray gene expression profiling of myometrium from non-pregnant and pregnant women. Mol Hum Reprod 9 (2003): 681-700.
- Azais H, Leroy A, Ghesquiere L, et al., Effects of adipokines and obesity on uterine contractility. Cytokine Growth Factor Rev 34 (2017): 59-66.
- AlSaif S, Mumtaz S, Wray S. A short review of adipokines, smooth muscle and uterine contractility. Life Sci 125 (2015): 2-8.
- Mumtaz S, AlSaif S, Wray S, et al., Inhibitory effect of visfatin and leptin on human and rat myometrial contractility. Life Sci 125 (2015): 57-62.
- Moynihan AT, Hehir MP, Glavey SV, et al., Inhibitory effect of leptin on human uterine contractility in vitro. Am J Obstet Gynecol 195 (2006): 504-509.
- Hehir MP, Morrison JJ. The adipokine apelin and human uterine contractility. Am J Obstet Gynecol 206 (2012): e351-355.
- Balogh O, Staub LP, Gram A, et al. Leptin in the canine uterus and placenta: possible implications in pregnancy. Reprod Biol Endocrinol 13 (2015): 13.
- Markowska A, Rucinski M, Drews K, et al., Further studies on leptin and leptin receptor expression in myometrium and uterine myomas. Eur J Gynaecol Oncol 26 (2005): 517-525.
- Dobrzyn K, Smolinska N, Kiezun M, et al., Adiponectin: A New Regulator of Female Reproductive System. Int J Endocrinol 2018 (2018): 7965071.
- Smolinska N, Maleszka A, Dobrzyn K, et al., Expression of adiponectin and adiponectin receptors 1 and 2 in the porcine uterus, conceptus, and trophoblast during early pregnancy. Theriogenology 82 (2014): 951-965.
- Annie L, Gurusubramanian G, Roy VK. Estrogen and progesterone dependent expression of visfatin/NAMPT regulates proliferation and apoptosis in mice uterus during estrous cycle. J Steroid Biochem Mol Biol 185 (2019): 225-236.
- O'Brien M, Earley P, Morrison JJ, et al., Ghrelin in the human myometrium. Reprod Biol Endocrinol 8 (2010): 55.
- Myers MG, Leibel RL, Seeley RJ, et al., Obesity and leptin resistance: distinguishing cause from effect. Trends Endocrinol Metab 21 (2010): 643-651.
- Asselin E, Drolet P, Fortier MA. Cellular mechanisms involved during oxytocin-induced prostaglandin F2alpha production in endometrial epithelial cells in vitro: role of cyclooxygenase-2. Endocrinology 138 (1997): 4798-4805.
- Erkinheimo TL, Saukkonen K, Narko K, et al., Expression of cyclooxygenase-2 and prostanoid receptors by human myometrium. J Clin Endocrinol Metab 85 (2000): 3468-3475.
- Slattery MM, Friel AM, Healy DG, et al., Uterine relaxant effects of cyclooxygenase-2 inhibitors in vitro. Obstet Gynecol 98 (2001): 563-569.
Supplementary Data
Supplementary Table 1: List of genes (and encoded proteins) associated with parturition, OXT signaling, and obesity with primers for real time-qPCR.
|
Gene |
Primers |
Encodes |
|
|
Adipoq |
Forward |
AGGGAGAGAAAGGAGATGCAG |
Adiponectin |
|
Reverse |
CTTTCCTGCCAGGGGTTC |
||
|
Avp |
Forward |
CTACGCTCTCCGCTTGTTTC |
Arginine vasopressin |
|
Reverse |
GGGCAGTTCTGGAAGTAGCA |
||
|
Avpr1 |
Forward |
GGGATACCAATTTCGTTTGG |
Arginine vasopressin receptor 1A |
|
Reverse |
AAGCCAGTAACGCCGTGAT |
||
|
Dbn1 |
Forward |
TAACCCACGGGAGTTCTTCA |
Debrin 1 |
|
Reverse |
AAAGGGCAGTACGGACGAC |
||
|
Edn1 |
Forward |
TCCTTGATGGACAAGGAGTGT |
Endothelin 1 |
|
Reverse |
CCCAGTCCATACGGTACGA |
||
|
Gapdh |
Forward |
GAGCCAAACGGGTCATCA |
Glyceraldehyde 3-phosphate dehydrogenase |
|
Reverse |
CATATTTCTCGTGGTTCACACC |
||
|
Ghrl |
Forward |
CCAGAGGACAGAGGACAAGC |
Ghrelin |
|
Reverse |
ACATCGAAGGGAGCATTGAA |
||
|
Gja1 |
Forward |
TTTGACTTCAGCCTCCAAGG |
Connexin 43 |
|
Reverse |
CATGTCTGGGCACCTCTCTT |
||
|
Gucy1a3 |
Forward |
TACACTCGCTTTGACCAGCA |
Guanylate cyclase soluble subunit α3 |
|
Reverse |
AATATGCATCCCCGATGGT |
||
|
Gucy1b3 |
Forward |
CGTCTCAAAGGCCAAATGAT |
Guanylate cyclase soluble subunit β3 |
|
Reverse |
TCGTCCAGGTTCATCACACT |
||
|
Kcnq1 |
Forward |
ACTGCTGACCCCCATCAC |
Potassium voltage-gated channel Q member 1 |
|
Reverse |
CATGCGCCTGATGACCTT |
||
|
Lep |
Forward |
CAGGATCAATGACATTTCACACA |
Leptin |
|
Reverse |
GCTGGTGAGGACCTGTTGAT |
||
|
Lpar1 |
Forward |
GCCTCTACTTCCAGCCCTGT |
Lysophosphatidic acid receptor 1 |
|
Reverse |
GCACTGTTGTTCGTTCATGG |
||
|
Mapk1 |
Forward |
AAGAACTCATTTTTGAAGAGACTGC |
MAP Kinase 1 |
|
Reverse |
CTCTGAGCCCTTGTCCTGA |
||
|
Mylk |
Forward |
GCCAGGTCACTATGACAGTCC |
Myosin light chain kinase |
|
Reverse |
CGTCGTGAAGCCAGATGAC |
||
|
Mypt3 |
Forward |
CCTCGGAAGCATGTCCTCT |
Myosin phosphatase target subunit 3 |
|
Reverse |
GGTAAGGAACTGGCGGACT |
||
|
Nampt |
Forward |
TGTTCCAGGCTATTCTGTTCC |
Visfatin |
|
Reverse |
TTCAAAAGCATCTTTCTCATGG |
||
|
Nppa |
Forward |
CAACACAGATCTGATGGATTTCA |
Natriuretic peptide A |
|
Reverse |
CCTCATCTTCTACCGGCATC |
||
|
Npr1 |
Forward |
TGGAGACACAGTCAACACAGC |
Natriuretic peptide receptor 1 |
|
Reverse |
CGAAGACAAGTGGATCCTGAG |
||
|
Oxtr |
Forward |
AGCGTCTGGGACGTCAAT |
Oxytocin receptor |
|
Reverse |
GTTGAGGCTGGCCAAGAG |
||
|
Oxt |
Forward |
CACCTACAGCGGATCTCAGAC |
Oxytocin |
|
Reverse |
CGAGGTCAGAGCCAGTAAGC |
||
|
Pdgfra |
Forward |
AAGACCTGGGCAAGAGGAAC |
Platelet-derived growth factor receptor alpha |
|
Reverse |
GAACCTGTCTCGATGGCACT |
||
|
Pgr |
Forward |
TGCACCTGATCTAATCCTAAATGA |
Progesterone receptor (PGR) |
|
Reverse |
GGTAAGGCACAGCGAGTAGAA |
||
|
Pgrmc1 |
Forward |
CACAGCAGGAGACCCTGAGT |
PGR membrane component 1 |
|
Reverse |
CTCCCCTTCCTTCAGCAGTT |
||
|
Pgrmc2 |
Forward |
TGTTCGAGAATGGGAAATGC |
PGR membrane component 2 |
|
Reverse |
GATCCTTGGTGTCCTCCTCA |
||
|
Plcb1 |
Forward |
TCGATGAGAAGCCCAAGC |
Phospholipase C beta 1 |
|
Reverse |
GGCAGCCTTTTGAACTTGTC |
||
|
Prkcab |
Forward |
TCAAGCCGGAAAACCTCTTA |
Protein kinase cAMP-activated catalytic beta |
|
Reverse |
CTTGACTCTTTTGGCGAACC |
||
|
Prkca |
Forward |
CAAGGGATGAAATGTGACACC |
Protein kinase C alpha |
|
Reverse |
CCTCTTCTCTGTGTGATCCATTC |
||
|
Prkcg |
Forward |
GTTTGAGGCCTGCAATTACC |
Protein kinase C gamma |
|
Reverse |
AGAGTTCGTCGGAGCCTCT |
||
|
Ptgs1 |
Forward |
CCTCTTTCCAGGAGCTCACA |
Cyclooxygenase (COX)-1 |
|
Reverse |
TCGATGTCACCGTACAGCTC |
||
|
Ptgs2 |
Forward |
GGGAGTCTGGAACATTGTGAA |
Cyclooxygenase (COX)-2 |
|
Reverse |
TGTCAATCAAATATGATCTGGATGT |
Supplementary Table 2: All data for experiment 1: Gene expression comparing groups (lean vs. obese) and treatment (± OXT).
|
Gene |
Lean |
Obese |
|||||
|
-OXT |
+OXT |
-OXT |
+OXT |
Main Effect |
|||
|
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
p-value for inter- actiona |
Comparisonsb |
p-value |
|
|
Adipoq |
0.556 (0.209, 1.484) |
0.167 (0.067, 0.414) |
0.683 (0.275, 1.693) |
0.203 (0.082, 0.504) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.6515 0.0110 |
|
Avp |
0.706 (0.296, 1.687) |
1.958 (0.874, 4.385) |
1.420 (0.594, 3.393) |
2.707 (1.208, 6.062) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.2228 0.0476 |
|
Avpr
|
0.941 (0.706, 1.253) |
1.156 (0.887, 1.507) |
0.688 (0.527, 0.896) |
0.627 (0.481, 0.817) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0016 0.7015 |
|
Dbn1 |
0.994 (0.872, 1.133) |
1.418 (1.256, 1.602) |
0.838 (0.742, 0.946) |
1.145 (1.014, 1.293) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0031 <.0001 |
|
Edn1
|
0.992 (0.882, 1.115) |
0.575 (0.516, 0.641) |
0.599 (0.537, 0.667) |
0.516 (0.463, 0.575) |
0.0012 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0569 <.0001 |
|
Ghrl |
0.982 (0.804, 1.200) |
1.545 (1.283, 1.861) |
0.846 (0.703, 1.019) |
1.268 (1.053, 1.527 |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0638 <.0001 |
|
Gja1 |
0.978 (0.769, 1.244) |
1.098 (0.879, 1.371) |
1.059 (0.848, 1.323) |
1.081 (0.865, 1.351) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.7807 0.5443 |
|
Gucy1a3
|
0.990 (0.852, 1.151) |
0.493 (0.429, 0.566) |
0.769 (0.669, 0.884) |
0.633 (0.551, 0.728) |
0.0013 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0523 0.0180 |
|
Gucy1b3
|
0.990 (0.861, 1.139) |
0.522 (0.459, 0.595) |
0.781 (0.686, 0.890) |
0.661 (0.581, 0.753) |
0.0012 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0725 0.0173 |
|
Kcnq1
|
0.986 (0.817, 1.190) |
0.622 (0.522, 0.740) |
0.581 (0.488, 0.692) |
0.538 (0.452, 0.641) |
0.0356 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0011 0.5257 0.0003 |
|
Lep |
0.571 (0.218, 1.493) |
0.396 (0.163, 0.965) |
1.513 (0.621, 3.687) |
1.096 (0.419, 2.867) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0323 0.4391 |
|
Lpar1 |
0.984 (0.852, 1.138) |
0.911 (0.797, 1.042) |
0.922 (0.806, 1.054) |
1.034 (0.904, 1.182) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.6149 0.7411 |
|
Mapk1
|
0.998 (0.912, 1.092) |
0.712 (0.655, 0.774) |
0.822 (0.756, 0.893) |
0.878 (0.808, 0.955) |
<.0001 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.2569 0.0033 |
|
Mylk |
0.952 (0.730, 1.243) |
0.395 (0.309, 0.506) |
0.582 (0.455, 0.745) |
0.465 (0.363, 0.595) |
0.0130 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.1958 0.0100 |
|
Mypt3
|
0.997 (0.897, 1.108) |
0.533 (0.483, 0.588) |
0.743 (0.674, 0.820) |
0.547 (0.496, 0.603) |
0.0031 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0001 0.0003 |
|
Nampt |
0.989 (0.763,1.283) |
0.676 (0.531,0.860) |
1.283 (1.009,1.632) |
0.865 (0.680,1.100) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0392 0.0027 |
|
Nppa |
0.994 (0.860, 1.150) |
0.797 (0.697, 0.912) |
0.890 (0.778, 1.019) |
0.717 (0.627, 0.821) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.1094 0.0026 |
|
Npr1 |
0.994 (0.826, 1.198) |
0.462 (0.389, 0.548) |
0.629 (0.530, 0.748) |
0.435 (0.366, 0.517) |
0.0281 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
<.0001 0.0047 0.0011 |
|
Oxt |
0.934 (0.512, 1.703) |
1.551 (0.889, 2.705) |
2.373 (1.361, 4.138) |
2.164 (1.241, 3.774) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0335 0.4840 |
|
Oxtr |
0.909 (0.683, 1.210) |
2.994 (2.299, 3.900) |
0.570 (0.438, 0.743) |
1.254 (0.963, 1.634) |
NS |
Obese vs Lean No Oxt vs Oxt |
<.0001 <.0001 |
|
Pdgfra |
0.986 (0.860, 1.131) |
0.956 (0.842, 1.085) |
0.867 (0.764, 0.984) |
0.792 (0.698, 0.899) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0157 0.3245 |
|
Pgr |
0.983 (0.816, 1.184) |
0.894 (0.753, 1.062) |
0.792 (0.667, 0.940) |
0.759 (0.639, 0.902) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0326 0.4272 |
|
Pgrmc1 |
0.994 (0.872, 1.132) |
0.720 (0.638, 0.812) |
0.868 (0.769, 0.980) |
0.855 (0.758, 0.965) |
0.0169 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0011 0.8576 0.1296 |
|
Pgrmc2
|
0.985 (0.792, 1.224) |
0.667 (0.545, 0.816) |
0.909 (0.743, 1.112) |
1.079 (0.882, 1.320) |
0.0097 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0124 0.2260 0.5820 |
|
Plcb1 |
0.975 (0.816, 1.165) |
1.645 (1.395, 1.940) |
0.798 (0.677, 0.941) |
1.516 (1.285, 1.788) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0980 <.0001 |
|
Prkacb |
0.990 (0.891, 1.100) |
0.716 (0.650, 0.790) |
0.811 (0.736, 0.894) |
0.707 (0.642, 0.780) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0553 0.0002 |
|
Prkca |
0.988 (0.856, 1.140) |
0.671 (0.587, 0.766) |
0.736 (0.644, 0.840 |
0.684 (0.599, 0.782) |
0.0251 |
Lean: Oxt vs No Oxt Obese: Oxt vs No Oxt No Oxt: Obese vs Lean |
0.0005 0.4358 0.0049 |
|
Prkcg |
0.994 (0.810, 1.219) |
1.152 (0.954, 1.392) |
0.615 (0.509, 0.743) |
0.783 (0.648, 0.946) |
NS |
Obese vs Lean No Oxt vs Oxt |
<.0001 0.0426 |
|
Ptgs2 |
0.930 (0.633, 1.367) |
0.687 (0.481, 0.981) |
0.830 (0.565, 1.220) |
0.672 (0.457, 0.987) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.7118 0.1584 |
|
Ptgs1 |
0.989 (0.829, 1.181) |
0.587 (0.499, 0.692) |
0.670 (0.569, 0.789) |
0.539 (0.457, 0.635) |
NS |
Obese vs Lean No Oxt vs Oxt |
0.0116 0.0003 |
|
ap<0.05 is significant bWhere interaction between group/BMI and OXT treatment (yes,no) was significant (p<0.05), pre-specified comparisons were examined within the ANOVA model. For these comparisons, a Bonferroni adjustment was used, such that p<0.0167 was considered significant. Where the interaction between group and OXT was not significant (NS), the interaction was removed and only the main effects of group and treatment (OXT) were examined. |
|||||||
Supplementary Table 3: All data for experiment 2: Gene expression comparing vehicle vs. regular- and high-dose OXT in obese uteri.
|
Veh (-OXT) |
Reg-dosea |
Hi-doseb |
||||
|
Gene |
Mean (95% CI) |
Mean (95% CI) |
Mean (95% CI) |
p-value for effect of OXTc |
Comparisond |
p-value |
|
Adipoq |
0.683 (0.317,1.472) |
0.203 (0.094,0.438) |
0.252 (0.101, 0.624) |
0.0723 |
||
|
Avp |
1.420 (0.632, 3.191) |
2.707 (1.279, 5.727) |
1.908 (0.786, 4.632) |
0.4753 |
||
|
Avpr |
0.688 (0.524, 0.902) |
0.627 (0.478, 0.823) |
0.716 (0.519, 0.987) |
0.7828 |
||
|
Dbn1 |
0.838 (0.724, 0.971) |
1.145 (0.989, 1.326) |
1.271 (1.068, 1.512) |
0.0026 |
Veh vs Hi |
0.0013 |
|
Reg vs Hi |
0.3446 |
|||||
|
Reg vs Veh |
0.0057 |
|||||
|
Edn1 |
0.599 (0.522, 0.687) |
0.516 (0.450, 0.592) |
0.543 (0.462, 0.639) |
0.2874 |
||
|
Ghrl |
0.846 (0.706, 1.014) |
1.268 (1.058, 1.520) |
1.246 (1.005, 1.544) |
0.0072 |
Veh vs Hi |
0.0100 |
|
Reg vs Hi |
0.8950 |
|||||
|
Reg vs Veh |
0.0041 |
|||||
|
Gja1 |
1.059 (0.807, 1.389) |
1.081 (0.824, 1.418) |
1.503 (1.091, 2.072) |
0.1870 |
||
|
Gucy1a3
|
0.769 (0.649, 0.911) |
0.633 (0.534, 0.750) |
0.492 (0.403, 0.601) |
0.0086 |
Veh vs Hi |
0.0024 |
|
Reg vs Hi |
0.0592 |
|||||
|
Reg vs Veh |
0.1038 |
|||||
|
Gucy1b3 |
0.781 (0.681, 0.897) |
0.661 (0.576, 0.759) |
0.618 (0.525, 0.727) |
0.0745 |
||
|
Kcnq1 |
0.581 (0.467, 0.723) |
0.538 (0.433, 0.670) |
0.445 (0.344, 0.576) |
0.2674 |
||
|
Lep |
1.513 (0.885, 2.587) |
1.096 (0.614, 1.955) |
0.961 (0.509, 1.812) |
0.4852 |
||
|
Lpar1 |
0.922 (0.795, 1.069) |
1.034 (0.892, 1.199) |
1.071 (0.899, 1.276) |
0.3439 |
||
|
Mapk1 |
0.822 (0.759, 0.890) |
0.878 (0.811, 0.951) |
0.827 (0.753, 0.909) |
0.4228 |
||
|
Mylk |
0.582 (0.441, 0.768) |
0.465 (0.352, 0.614) |
0.428 (0.308, 0.595) |
0.2963 |
||
|
Mypt3 |
0.743 (0.657, 0.841) |
0.547 (0.483, 0.619) |
0.530 (0.458, 0.613) |
0.0018 |
Veh vs Hi |
0.0018 |
|
Reg vs Hi |
0.7351 |
|||||
|
Reg vs Veh |
0.0018 |
|||||
|
Nampt |
1.283 (0.963, 1.709) |
0.865 (0.650, 1.152) |
1.076 (0.766, 1.510) |
0.1511 |
||
|
Nppa |
0.890 (0.758, 1.046) |
0.717 (0.611, 0.842) |
0.761 (0.629, 0.921) |
0.1502 |
||
|
Npr1 |
0.629 (0.501, 0.791) |
0.435 (0.346, 0.547) |
0.412 (0.314, 0.540) |
0.0339 |
Veh vs Hi |
0.0221 |
|
Reg vs Hi |
0.7446 |
|||||
|
Reg vs Veh |
0.0281 |
|||||
|
Oxt |
2.373 (1.305, 4.313) |
2.164 (1.191, 3.934) |
1.707 (0.842, 3.462) |
0.7503 |
||
|
Oxtr |
0.570 (0.431, 0.755) |
1.254 (0.948, 1.660) |
1.346 (0.966, 1.876) |
0.0006 |
Veh vs Hi |
0.0007 |
|
Reg vs Hi |
0.7338 |
|||||
|
Reg vs Veh |
0.0007 |
|||||
|
Pdgfra |
0.867 (0.752, 1.000) |
0.792 (0.687, 0.913) |
0.878 (0.742, 1.039) |
0.5343 |
||
|
Pgr |
0.792 (0.661, 0.949) |
0.759 (0.634, 0.910) |
0.706 (0.570, 0.875) |
0.6914 |
||
|
Pgrmc1 |
0.868 (0.752, 1.002) |
0.855 (0.740, 0.988) |
0.949 (0.800, 1.125) |
0.5897 |
||
|
Pgrmc2 |
0.909 (0.710, 1.164) |
1.079 (0.842, 1.382) |
1.127 (0.841, 1.511) |
0.4410 |
||
|
Plcb1 |
0.798 (0.672, 0.948) |
1.516 (1.277, 1.800) |
1.180 (0.963, 1.446) |
0.0002 |
Veh vs Hi |
0.0067 |
|
Reg vs Hi |
0.0633 |
|||||
|
Reg vs Veh |
<0.0001 |
|||||
|
Prkacb |
0.811 (0.740, 0.888) |
0.707 (0.646, 0.775) |
0.731 (0.656, 0.814) |
0.0974 |
||
|
Prkca |
0.736 (0.627, 0.863) |
0.684 (0.584, 0.803) |
0.600 (0.497, 0.725) |
0.2462 |
||
|
Prkcg |
0.615 (0.523, 0.723) |
0.783 (0.666, 0.921) |
0.924 (0.763, 1.120) |
0.0103 |
Veh vs Hi |
0.0034 |
|
Reg vs Hi |
0.1821 |
|||||
|
Reg vs Veh |
0.0402 |
|||||
|
Ptgs1 |
0.670 (0.531, 0.845) |
0.539 (0.427, 0.679) |
0.560 (0.426, 0.737) |
0.3563 |
||
|
Ptgs2 |
0.830 (0.533, 1.295) |
0.672 (0.431, 1.047) |
1.726 (1.061, 2.807) |
0.0220 |
Veh vs Hi |
0.0319 |
|
Reg vs Hi |
0.0083 |
|||||
|
Reg vs Veh |
0.4808 |
|||||
|
aHi-dose=high-dose OXT bReg-dose=regular dose OXT cp<0.05 is significant dWhere there was a significant effect of OXT (p<0.05), pairwise comparisons were carried out within the ANOVA model. A Bonferroni adjustment was used, such that p<0.0167 was considered significant for these comparisons. |
||||||
